9ICW
 
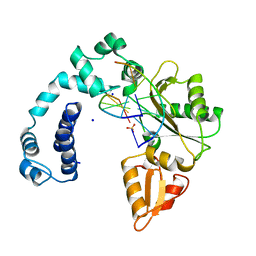 | | DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; NATIVE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*CP*TP*GP*T)-3'), PROTEIN (DNA POLYMERASE BETA (E.C.2.7.7.7)), ... | | Authors: | Pelletier, H, Sawaya, M.R. | | Deposit date: | 1995-12-16 | | Release date: | 1996-11-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity
Biochemistry, 35, 1996
|
|
9ICS
 
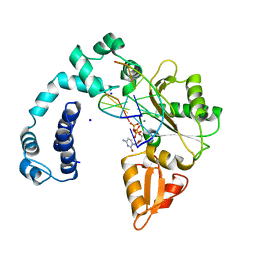 | | DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2',3'-DIDEOXYCYTIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DDCTP AND MNCL2 | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*AP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*CP*TP*GP*T)-3'), ... | | Authors: | Pelletier, H, Sawaya, M.R. | | Deposit date: | 1995-12-16 | | Release date: | 1996-11-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase beta
Biochemistry, 35, 1996
|
|
9ICP
 
 | |
9ICN
 
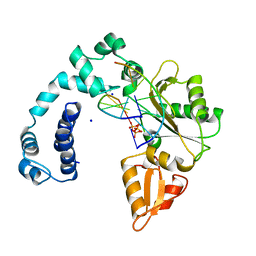 | | DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2',3'-DIDEOXYCYTIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DDCTP AND MGCL2 | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*AP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*CP*TP*GP*T)-3'), ... | | Authors: | Pelletier, H, Sawaya, M.R. | | Deposit date: | 1995-12-16 | | Release date: | 1995-12-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase beta
Biochemistry, 35, 1996
|
|
9ICK
 
 | |
9ICQ
 
 | |
2Z9H
 
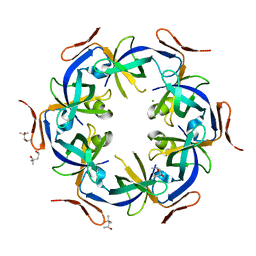 | | Ethanolamine utilization protein, EutN | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CHLORIDE ION, Ethanolamine utilization protein eutN | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The crystal structure of ethanolamine utilization protein EutN from E. coli
To be Published
|
|
2Y3L
 
 | | Structure of segment MVGGVVIA from the amyloid-beta peptide (Ab, residues 35-42), alternate polymorph 2 | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y5Y
 
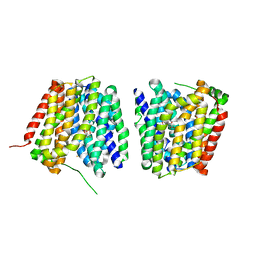 | | Crystal structure of LacY in complex with an affinity inactivator | | Descriptor: | 2-sulfanylethyl beta-D-galactopyranoside, BARIUM ION, LACTOSE PERMEASE | | Authors: | Chaptal, V, Kwon, S, Sawaya, M.R, Guan, L, Kaback, H.R, Abramson, J. | | Deposit date: | 2011-01-19 | | Release date: | 2011-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Crystal Structure of Lactose Permease in Complex with an Affinity Inactivator Yields Unique Insight Into Sugar Recognition.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y2A
 
 | | Structure of segment KLVFFA from the amyloid-beta peptide (Ab, residues 16-21), alternate polymorph I | | Descriptor: | ACETATE ION, AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y3J
 
 | | Structure of segment AIIGLM from the amyloid-beta peptide (Ab, residues 30-35) | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y3K
 
 | | Structure of segment MVGGVVIA from the amyloid-beta peptide (Ab, residues 35-42), alternate polymorph 1 | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y29
 
 | | Structure of segment KLVFFA from the amyloid-beta peptide (Ab, residues 16-21), alternate polymorph III | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1ZUR
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R1F) | | Descriptor: | CHLORIDE ION, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-4-phenyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | Deposit date: | 2005-05-31 | | Release date: | 2006-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of spin labeled T4 Lysozyme (V131R1F)
To be Published
|
|
7N8R
 
 | | FGTGFG segment from the Nucleoporin p54, residues 63-68 | | Descriptor: | FGTGFG segment from the Nucleoporin p54, residues 63-68, trifluoroacetic acid | | Authors: | Hughes, M.P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Extended beta-Strands Contribute to Reversible Amyloid Formation.
Acs Nano, 16, 2022
|
|
7N2H
 
 | | X-Ray structure of a sequence variant of a repeat segment of the yeast prion New1p | | Descriptor: | prion New1p | | Authors: | Richards, L.S, Flores, M.D, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.102 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2G
 
 | | MicroED structure of human CPEB3 segment(154-161) kinked polymorph phased by ARCIMBOLDO-BORGES | | Descriptor: | CPEB3 | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.201 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2I
 
 | | MicroED structure of human LECT2 (45-53) phased by ARCIMBOLDO-BORGES | | Descriptor: | LECT2 | | Authors: | Richards, L.S, Flores, M.D, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.402 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2L
 
 | | MicroED structure of a mutant mammalian prion segment | | Descriptor: | prion protein | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2D
 
 | | MicroED structure of human zinc finger protein 292 segment (534-542) phased by ARCIMBOLDO-BORGES | | Descriptor: | DIMETHYL SULFOXIDE, zinc finger protein 292 | | Authors: | Richards, L.S, Flores, M.D, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.503 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2J
 
 | | MicroED structure of a mutant mammalian prion segment phased by ARCIMBOLDO-BORGES | | Descriptor: | prion protein | | Authors: | Richards, L.S, Flores, M.D, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.5 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2E
 
 | | MicroED structure of human CPEB3 segment (154-161) straight polymorph | | Descriptor: | CPEB3 | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2K
 
 | | MicroED structure of sequence variant of repeat segment of the yeast prion New1p phased by ARCIMBOLDO-BORGES | | Descriptor: | prion New1p | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.301 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2F
 
 | | MicroED structure of human CPEB3 segment (154-161) straight polymorph phased by ARCIMBOLDO-BORGES | | Descriptor: | CPEB3 | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
6AVB
 
 | | CryoEM structure of Mical Oxidized Actin (Class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle | | Authors: | Grintsevich, E.E, Ge, P, Sawaya, M.R, Yesilyurt, H.G, Terman, J.R, Zhou, Z.H, Reisler, E. | | Deposit date: | 2017-09-01 | | Release date: | 2018-01-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Catastrophic disassembly of actin filaments via Mical-mediated oxidation.
Nat Commun, 8, 2017
|
|
