7Q58
 
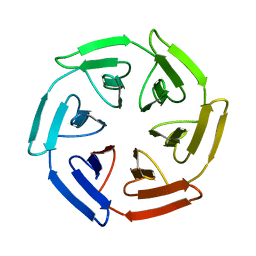 | | Crystal structure of the SAKe6BR designer protein | | Descriptor: | SAKe6BR | | Authors: | Wouters, S.M.L, Noguchi, H, Velupla, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-11-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
6NPR
 
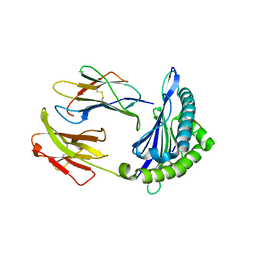 | | Crystal structure of H-2Dd with C84-C139 disulfide in complex with gp120 derived peptide P18-I10 | | Descriptor: | ARG-GLY-PRO-GLY-ARG-ALA-PHE-VAL-THR-ILE, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Toor, J, McShan, A.C, Tripathi, S.M, Sgourakis, N.G. | | Deposit date: | 2019-01-18 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Molecular determinants of chaperone interactions on MHC-I for folding and antigen repertoire selection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6NBL
 
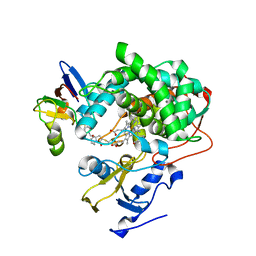 | | Cytochrome P450cam-putidaredoxin complex bound to camphor and cyanide | | Descriptor: | 1,1'-hexane-1,6-diyldipyrrolidine-2,5-dione, CALCIUM ION, CAMPHOR, ... | | Authors: | Follmer, A.H, Tripathi, S.M, Poulos, T.L. | | Deposit date: | 2018-12-07 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand and Redox Partner Binding Generates a New Conformational State in Cytochrome P450cam (CYP101A1).
J. Am. Chem. Soc., 141, 2019
|
|
6OF7
 
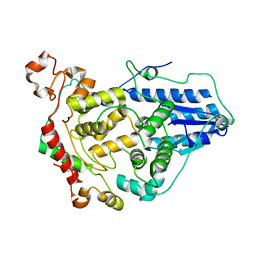 | | Crystal structure of the CRY1-PER2 complex | | Descriptor: | Cryptochrome-1, Period circadian protein homolog 2 | | Authors: | Michael, A.K, Fribourgh, J.L, Tripathi, S.M, Partch, C.L. | | Deposit date: | 2019-03-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Dynamics at the serine loop underlie differential affinity of cryptochromes for CLOCK:BMAL1 to control circadian timing.
Elife, 9, 2020
|
|
6X61
 
 | |
6L76
 
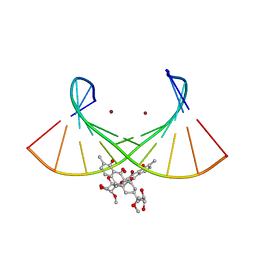 | | Crystal structure of the Ni(II)(Chro)2-d(TTGGGCCGAA/TTCGGCCCAA) complex at 2.94 angstrom resolution | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Hou, M.H, Jhan, C.R, Satange, R.B, Lin, S.M. | | Deposit date: | 2019-10-31 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Targeting the ALS/FTD-associated A-DNA kink with anthracene-based metal complex causes DNA backbone straightening and groove contraction.
Nucleic Acids Res., 49, 2021
|
|
6UYK
 
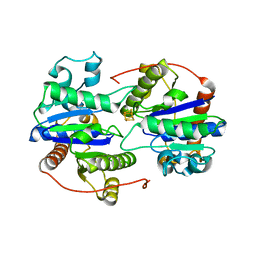 | | Dark-operative protochlorophyllide oxidoreductase in the nucleotide-free form. | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase iron-sulfur ATP-binding protein | | Authors: | Bacik, J.P, Imran, S.M.S, Watkins, M.B, Corless, E, Antony, E, Ando, N. | | Deposit date: | 2019-11-13 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The flexible N-terminus of BchL autoinhibits activity through interaction with its [4Fe-4S] cluster and released upon ATP binding.
J.Biol.Chem., 296, 2020
|
|
6EQI
 
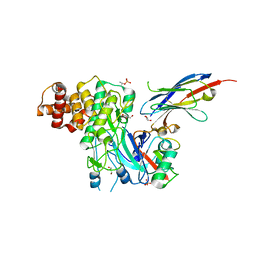 | | Structure of PINK1 bound to ubiquitin | | Descriptor: | GLYCEROL, Nb696, Serine/threonine-protein kinase PINK1, ... | | Authors: | Schubert, A.F, Gladkova, C, Pardon, E, Wagstaff, J.L, Freund, S.M.V, Steyaert, J, Maslen, S, Komander, D. | | Deposit date: | 2017-10-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of PINK1 in complex with its substrate ubiquitin.
Nature, 552, 2017
|
|
6PXN
 
 | |
6XHZ
 
 | | Alpha-lytic protease homolog N4 | | Descriptor: | N4: hypothetical protein, SULFATE ION | | Authors: | Nixon, C.F, Marqusee, S.M, Gee, C.L. | | Deposit date: | 2020-06-19 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploring the Evolutionary History of Kinetic Stability in the alpha-Lytic Protease Family.
Biochemistry, 60, 2021
|
|
7QKS
 
 | |
7QKR
 
 | | Cryo-EM structure of ABC transporter STE6-2p from Pichia pastoris with Verapamil at 3.2 A resolution | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, Dexverapamil, MAGNESIUM ION, ... | | Authors: | Schleker, E.S.M, Reinhart, C. | | Deposit date: | 2021-12-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional investigation of ABC transporter STE6-2p from Pichia pastoris reveals unexpected interaction with sterol molecules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6FK2
 
 | |
6TT5
 
 | | Crystal structure of DCLRE1C/Artemis | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Protein artemis, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
6TL1
 
 | | Crystal structure of the TASOR pseudo-PARP domain | | Descriptor: | GLYCEROL, Protein TASOR | | Authors: | Douse, C.H, Timms, R.T, Freund, S.M.V, Modis, Y. | | Deposit date: | 2019-11-29 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control.
Nat Commun, 11, 2020
|
|
6FPP
 
 | | Structure of S. pombe Mmi1 | | Descriptor: | GLYCEROL, MAGNESIUM ION, YTH domain-containing protein mmi1 | | Authors: | Stowell, J.A.W, Hill, C.H, Yu, M, Wagstaff, J.L, McLaughlin, S.H, Freund, S.M.V, Passmore, L.A. | | Deposit date: | 2018-02-11 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A low-complexity region in the YTH domain protein Mmi1 enhances RNA binding.
J. Biol. Chem., 293, 2018
|
|
4WFZ
 
 | |
4WFY
 
 | |
5E13
 
 | | Crystal structure of Eosinophil-derived neurotoxin in complex with the triazole double-headed ribonucleoside 11c | | Descriptor: | 3'-{4-[(4-amino-2-oxopyrimidin-1(2H)-yl)methyl]-1H-1,2,3-triazol-1-yl}-3'-deoxyadenosine, Non-secretory ribonuclease | | Authors: | Chatzileontiadou, D.S.M, Stravodimos, G.A, Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Triazole double-headed ribonucleosides as inhibitors of eosinophil derived neurotoxin.
Bioorg.Chem., 63, 2015
|
|
1GO1
 
 | |
1H1J
 
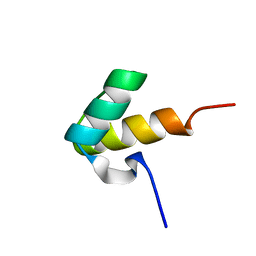 | |
4WFX
 
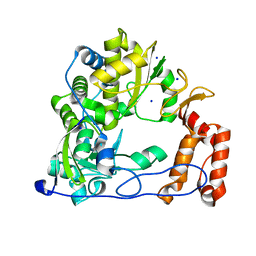 | |
5EQO
 
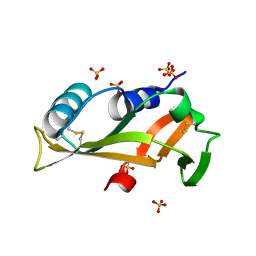 | |
8GU0
 
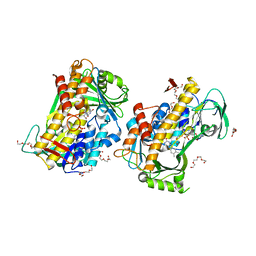 | | Crystal structure of a fungal halogenase RadH | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Non-heme halogenase radH, ... | | Authors: | Jiang, S.M, Brown, C.J. | | Deposit date: | 2022-09-09 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Further Characterization of Fungal Halogenase RadH and Its Homologs.
Biomolecules, 13, 2023
|
|
8GN3
 
 | | The crystal structure of ZBTB10 ZF1-2 in complex with telomeric vairant repeat TTGGGG | | Descriptor: | DNA (5'-D(*AP*TP*AP*CP*AP*AP*CP*CP*CP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*GP*GP*GP*TP*TP*GP*TP*A)-3'), ZINC ION, ... | | Authors: | Li, F.D, Wang, S.M. | | Deposit date: | 2022-08-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the recognition of telomeric variant repeat TTGGGG by broad-complex, tramtrack and bric-a-brac - zinc finger protein ZBTB10.
J.Biol.Chem., 299, 2023
|
|
