8V86
 
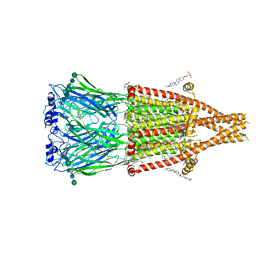 | | Alpha7-nicotinic acetylcholine receptor bound to GAT107 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (3aR,4S,9bS)-4-(4-bromophenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-12-04 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
1AB7
 
 | |
8UU6
 
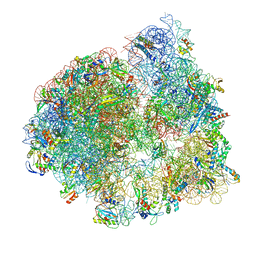 | | Cryo-EM structure of the ratcheted Listeria innocua 70S ribosome in complex with p/E-tRNA (structure II-A) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UUA
 
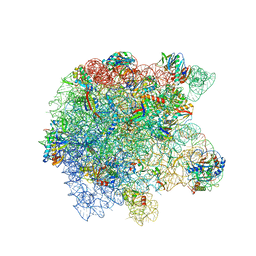 | | Cryo-EM structure of the Listeria innocua 50S ribosomal subunit in complex with HflXr (structure III) | | Descriptor: | 23S Ribosomal RNA, 5S Ribosomal RNA, GTPase HflX, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UU7
 
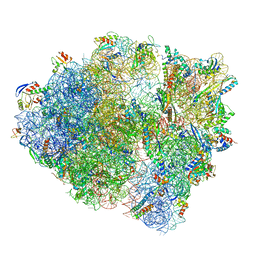 | | Cryo-EM structure of the Listeria innocua 70S ribosome in complex with HflXr, HPF, and E-site tRNA (structure II-B) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UU4
 
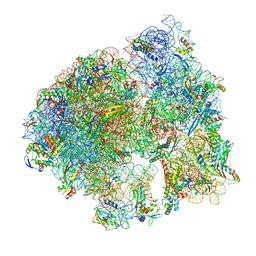 | | Cryo-EM structure of the Listeria innocua 70S ribosome in complex with HPF (structure I-A) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8WMJ
 
 | | structure of PSI-11CAC complex at Logrithmic growth phase | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
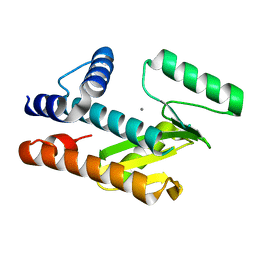
8VGZ
 
 | |
8VXQ
 
 | |
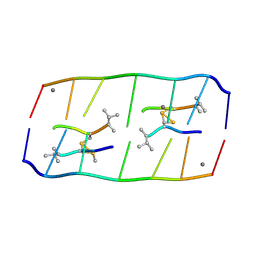
8XP8
 
 | |
8WM6
 
 | | The structure of PSI-CAC(L-14)of R.salina at 2.7 angstroms resolution | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
8XP9
 
 | |
8XPA
 
 | |
2L44
 
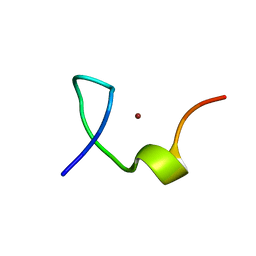 | | C-terminal zinc knuckle of the HIVNCp7 | | Descriptor: | C-terminal Zinc Knucle of the HIV-NCP7, ZINC ION | | Authors: | Quintal, S.M.O, Viegas, A.J.M, Cabrita, E.J, Farrell, N.P, Erhardt, S. | | Deposit date: | 2010-10-01 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Platinated DNA affects zinc finger conformation. Interaction of a platinated single-stranded oligonucleotide and the C-terminal zinc finger of nucleocapsid protein HIVNCp7.
Biochemistry, 51, 2012
|
|
2L9N
 
 | |
6Y5V
 
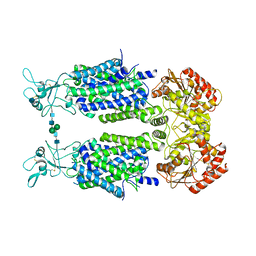 | | Structure of Human Potassium Chloride Transporter KCC3b (S45D/T940D/T997D) in KCl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L. | | Deposit date: | 2020-02-25 | | Release date: | 2020-07-15 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
6Y7F
 
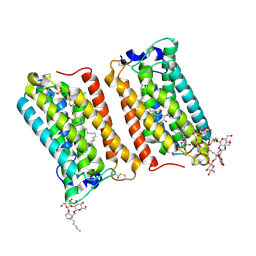 | | Crystal structure of human ELOVL fatty acid elongase 7 (ELOVL7) | | Descriptor: | CHLORIDE ION, Elongation of very long chain fatty acids protein 7, Octyl Glucose Neopentyl Glycol, ... | | Authors: | Nie, L, Pike, A.C.W, Bushell, S.R, Chu, A, Cole, V, Speedman, D, Rodstrom, K.E.J, Kupinska, K, Shrestha, L, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-28 | | Release date: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal structure of human ELOVL fatty acid elongase 7 (ELOVL7)
TO BE PUBLISHED
|
|
6Y5R
 
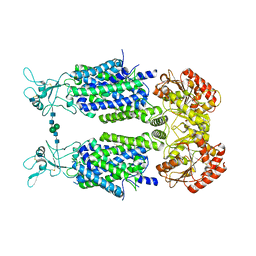 | | Structure of Human Potassium Chloride Transporter KCC3 S45D/T940D/T997D in NaCl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, B, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-25 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structure of Human Potassium Chloride Transporter KCC3 in NaCl
To be published
|
|
1BTA
 
 | |
2MZZ
 
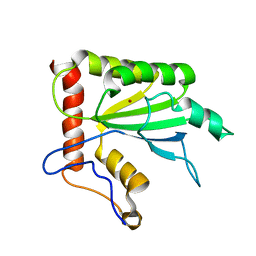 | | NMR structure of APOBEC3G NTD variant, sNTD | | Descriptor: | Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G variant, ZINC ION | | Authors: | Kouno, T, Luengas, E.M, Shigematu, M, Shandilya, S.M.D, Zhang, J, Chen, L, Hara, M, Schiffer, C.A, Harris, R.S, Matsuo, H. | | Deposit date: | 2015-02-28 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Vif-binding domain of the antiviral enzyme APOBEC3G.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1BTB
 
 | |
6PXP
 
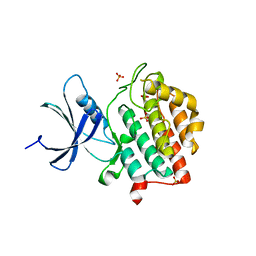 | | Human Casein Kinase 1 delta Site 2 mutant (K171E) | | Descriptor: | Casein kinase I isoform delta, SULFATE ION | | Authors: | Yee, L, Philpott, J.M, Tripathi, S.M, Partch, C.L. | | Deposit date: | 2019-07-26 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Casein kinase 1 dynamics underlie substrate selectivity and the PER2 circadian phosphoswitch.
Elife, 9, 2020
|
|
6SWG
 
 | |
6CWR
 
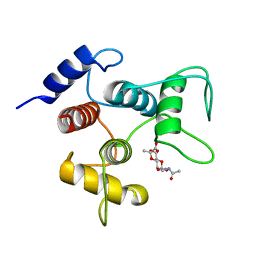 | | Crystal structure of SpaA-SLH/G46A/G109A in complex with 4,6-Pyr-beta-D-ManNAcOMe | | Descriptor: | Surface (S-) layer glycoprotein, methyl 2-(acetylamino)-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranoside | | Authors: | Blackler, R.J, Gagnon, S.M.L, Haji-Ghassemi, O, Evans, S.V. | | Deposit date: | 2018-03-30 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural basis of cell wall anchoring by SLH domains in Paenibacillus alvei.
Nat Commun, 9, 2018
|
|
7PVP
 
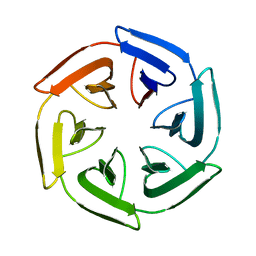 | | Crystal structure of the SAKe6BC designer protein | | Descriptor: | SAKe6BC | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-10-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
