2J0O
 
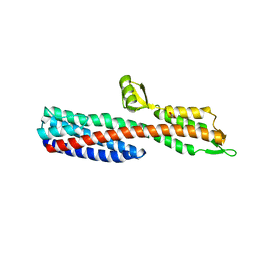 | | Shigella Flexneri IpaD | | Descriptor: | GLYCEROL, INVASIN IPAD | | Authors: | Johnson, S, Roversi, P, Lea, S.M. | | Deposit date: | 2006-08-04 | | Release date: | 2006-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD.
J. Biol. Chem., 282, 2007
|
|
1K24
 
 | |
2IXR
 
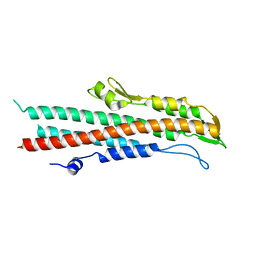 | | BipD of Burkholderia Pseudomallei | | Descriptor: | MEMBRANE ANTIGEN | | Authors: | Roversi, P, Johnson, S, Field, T, Deane, J.E, Galyov, E.E, Lea, S.M. | | Deposit date: | 2006-07-10 | | Release date: | 2006-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Self-Chaperoning of the Type III Secretion System Needle Tip Proteins Ipad and Bipd.
J.Biol.Chem., 282, 2007
|
|
2J0N
 
 | |
5XD1
 
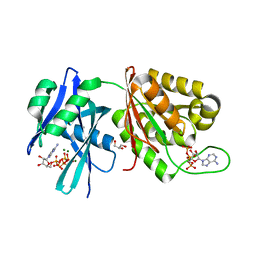 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with Ap5A, ATP and magnesium | | Descriptor: | ADENOSINE-5'-PENTAPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
J. Struct. Biol., 199, 2017
|
|
5XD3
 
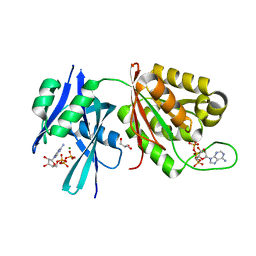 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with ATP (I) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
J. Struct. Biol., 199, 2017
|
|
5Y0A
 
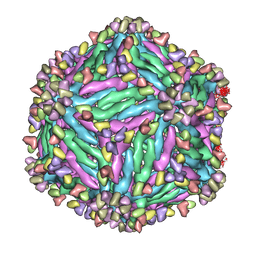 | |
5XD5
 
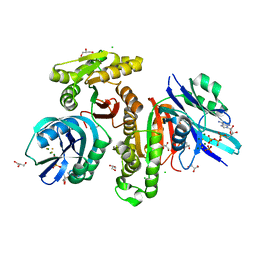 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with ATP, magnesium fluoride and phosphate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, FLUORIDE ION, ... | | Authors: | Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
J. Struct. Biol., 199, 2017
|
|
1LLS
 
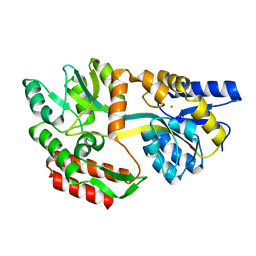 | | CRYSTAL STRUCTURE OF UNLIGANDED MALTOSE BINDING PROTEIN WITH XENON | | Descriptor: | Maltose-binding periplasmic protein, XENON | | Authors: | Rubin, S.M, Lee, S.-Y, Ruiz, E.J, Pines, A, Wemmer, D.E. | | Deposit date: | 2002-04-30 | | Release date: | 2002-09-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DETECTION AND CHARACTERIZATION OF XENON-BINDING SITES IN PROTEINS BY 129XE NMR SPECTROSCOPY
J.MOL.BIOL., 322, 2002
|
|
5XD4
 
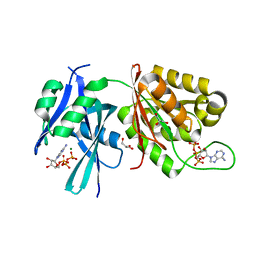 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with ATP (II) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
J. Struct. Biol., 199, 2017
|
|
5TUU
 
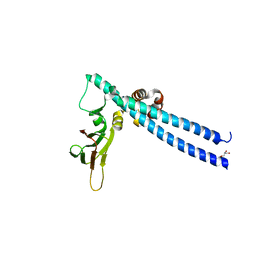 | |
5X3T
 
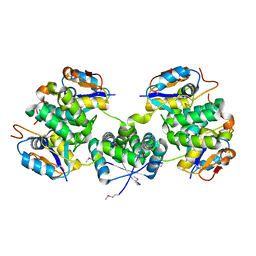 | | VapBC from Mycobacterium tuberculosis | | Descriptor: | Antitoxin VapB26, MAGNESIUM ION, Ribonuclease VapC26 | | Authors: | Kang, S.M, Kim, D.H, Yoon, H.J, Lee, B.J. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-07 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional details of the Mycobacterium tuberculosis VapBC26 toxin-antitoxin system based on a structural study: insights into unique binding and antibiotic peptides.
Nucleic Acids Res., 45, 2017
|
|
1M9L
 
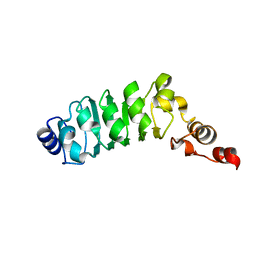 | | Relaxation-based Refined Structure Of Chlamydomonas Outer Arm Dynein Light Chain 1 | | Descriptor: | Outer Arm Dynein Light Chain 1 | | Authors: | Wu, H.W, Maciejewski, M.W, Marintchev, A, Benashski, S.E, Mullen, G.P, King, S.M. | | Deposit date: | 2002-07-29 | | Release date: | 2003-03-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Relaxation-based structure refinement and backbone molecular dynamics of the
Dynein motor domain-associated light chain
Biochemistry, 42, 2003
|
|
5XTU
 
 | | Crystal Structure of GDSL Esterase of Photobacterium sp. J15 | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Mazlan, S.N.H.S, Jonet, M.A, Leow, T.C, Ali, M.S.M, Rahman, R.N.Z.R.A. | | Deposit date: | 2017-06-21 | | Release date: | 2018-10-10 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystallization and structure elucidation of GDSL esterase of Photobacterium sp. J15.
Int. J. Biol. Macromol., 119, 2018
|
|
5XD2
 
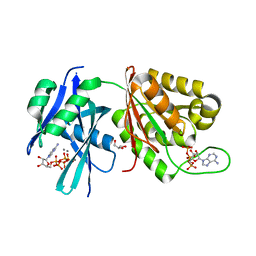 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with Ap5A, ATP and manganese | | Descriptor: | ADENOSINE-5'-PENTAPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
J. Struct. Biol., 199, 2017
|
|
1MJW
 
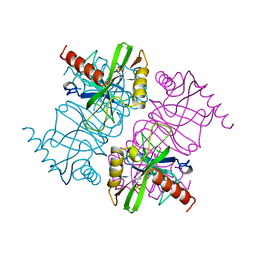 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE MUTANT D42N | | Descriptor: | INORGANIC PYROPHOSPHATASE, SULFATE ION | | Authors: | Oganesyan, V, Harutyunyan, E.H, Avaeva, S.M, Samygina, V.R, Huber, R. | | Deposit date: | 1997-02-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-dimensional structures of mutant forms of E. coli inorganic pyrophosphatase with Asp-->Asn single substitution in positions 42, 65, 70, and 97.
Biochemistry Mosc., 63, 1998
|
|
1MJX
 
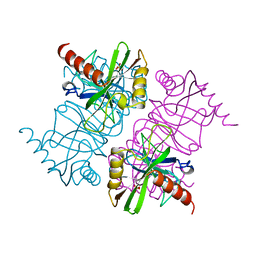 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE MUTANT D65N | | Descriptor: | INORGANIC PYROPHOSPHATASE, SULFATE ION | | Authors: | Oganesyan, V, Harutyunyan, E.H, Avaeva, S.M, Huber, R. | | Deposit date: | 1997-02-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-dimensional structures of mutant forms of E. coli inorganic pyrophosphatase with Asp-->Asn single substitution in positions 42, 65, 70, and 97.
Biochemistry Mosc., 63, 1998
|
|
1MJY
 
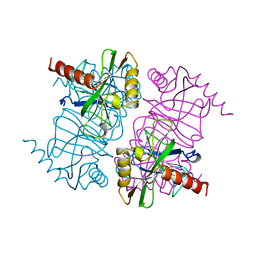 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE MUTANT D70N | | Descriptor: | INORGANIC PYROPHOSPHATASE | | Authors: | Oganesyan, V, Harutyunyan, E.H, Avaeva, S.M, Huber, R. | | Deposit date: | 1997-02-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structures of mutant forms of E. coli inorganic pyrophosphatase with Asp-->Asn single substitution in positions 42, 65, 70, and 97.
Biochemistry Mosc., 63, 1998
|
|
1KTA
 
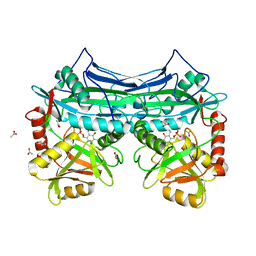 | | HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE : THREE DIMENSIONAL STRUCTURE OF THE ENZYME IN ITS PYRIDOXAMINE PHOSPHATE FORM. | | Descriptor: | 3-METHYL-2-OXOBUTANOIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ACETIC ACID, ... | | Authors: | Yennawar, N.H, Conway, M.E, Yennawar, H.P, Farber, G.K, Hutson, S.M. | | Deposit date: | 2002-01-15 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human mitochondrial branched chain aminotransferase reaction intermediates: ketimine and pyridoxamine phosphate forms
Biochemistry, 41, 2002
|
|
5VKY
 
 | |
5UQ2
 
 | |
4YPD
 
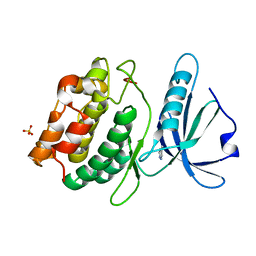 | | Crystal Structure of DAPK1 catalytic domain in complex with the hinge binding fragment 4-methylpyridazine | | Descriptor: | 4-methylpyridazine, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Grum-Tokars, V.L, Minasov, G, Roy, S.M, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of DAPK1 catalytic domain in complex with hinge binding fragments
To Be Published
|
|
6VV6
 
 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with JEB113 | | Descriptor: | 1-(4-fluorophenyl)-5-[3-(1H-indol-3-yl)propoxy]-1H-pyrazole-4-carboxylic acid, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Dias, M.V.B, Chaves-Pacheco, S.M. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.235 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
5TUV
 
 | |
1P6P
 
 | | Crystal Structure of Toad Liver Basic Fatty Acid-Binding Protein | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Di Pietro, S.M, Corsico, B, Perduca, M, Monaco, H.L, Santome, J.A. | | Deposit date: | 2003-04-30 | | Release date: | 2003-06-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Characterization of Toad Liver Basic Fatty Acid-Binding Protein
Biochemistry, 42, 2003
|
|
