4INE
 
 | |
2LO1
 
 | | NMR structure of the protein BC008182, a DNAJ-like domain from Homo sapiens | | Descriptor: | DnaJ homolog subfamily A member 1 | | Authors: | Dutta, S.K, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2012-01-09 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein BC008182, DNAJ homolog from Homo sapiens
To be Published
|
|
4ILK
 
 | | Crystal structure of short chain alcohol dehydrogenase (rspB) from E. coli CFT073 (EFI TARGET EFI-506413) complexed with cofactor NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MANGANESE (II) ION, Starvation sensing protein rspB, ... | | Authors: | Lukk, T, Wichelecki, D, Imker, H.J, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2012-12-31 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Crystal structure of short chain alcohol dehydrogenase (rspB) from E. coli CFT073 (EFI TARGET EFI-506413) complexed with cofactor NADH
To be Published
|
|
2LL0
 
 | | NMR structure of the putative ATPase regulatory protein YP_916642.1 from Paracoccus denitrificans | | Descriptor: | Uncharacterized protein | | Authors: | Serrano, P, Geralt, M, Wuthrich, K, Morales-Rios, E, Zarco-Zavala, M, Garcia-Trejo, J.J, Dutta, S.K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the putative ATPase regulatory protein YP_916642.1 from Paracoccus denitrificans
To be Published
|
|
2LRG
 
 | |
2LMI
 
 | |
2LHA
 
 | | Solution structure of C2B with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-1 | | Authors: | Joung, M, Mohan, S.K, Yu, C. | | Deposit date: | 2011-08-08 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Level Interaction of Inositol Hexaphosphate with the C2B Domain of Human Synaptotagmin I
Biochemistry, 2012
|
|
4IV8
 
 | |
3E9M
 
 | |
4IV0
 
 | |
2MHG
 
 | |
2M51
 
 | |
2LZ0
 
 | |
2MFN
 
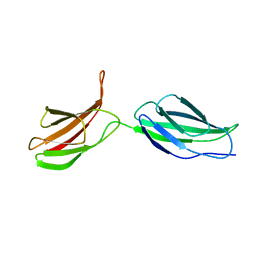 | | SOLUTION NMR STRUCTURE OF LINKED CELL ATTACHMENT MODULES OF MOUSE FIBRONECTIN CONTAINING THE RGD AND SYNERGY REGIONS, 10 STRUCTURES | | Descriptor: | FIBRONECTIN | | Authors: | Copie, V, Tomita, Y, Akiyama, S.K, Aota, S, Yamada, K.M, Venable, R.M, Pastor, R.W, Krueger, S, Torchia, D.A. | | Deposit date: | 1998-02-11 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of linked cell attachment modules of mouse fibronectin containing the RGD and synergy regions: comparison with the human fibronectin crystal structure.
J.Mol.Biol., 277, 1998
|
|
2MHE
 
 | |
2M52
 
 | |
2M7S
 
 | | NMR structure of RNA recognition motif 2 (RRM2) of Homo sapiens splicing factor, arginine/serine-rich 1 | | Descriptor: | Serine/arginine-rich splicing factor 1 | | Authors: | Dutta, S.K, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG), Partnership for T-Cell Biology (TCELL) | | Deposit date: | 2013-04-29 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of RNA recognition motif 2 (RRM2) of Homo sapiens splicing factor, arginine/serine-rich 1
To be Published
|
|
2MDZ
 
 | | NMR structure of the Paracoccus denitrificans Z-subunit determined in the presence of ADP | | Descriptor: | Uncharacterized protein | | Authors: | Serrano, P, Geralt, M, Wuthrich, K, Morales-Rios, E, Zarco-Zavala, M, Garcia-Trejo, J.J, Dutta, S.K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the putative ATPase regulatory protein YP_916642.1 from Paracoccus denitrificans
To be Published
|
|
4K3C
 
 | | The crystal structure of BamA from Haemophilus ducreyi lacking POTRA domains 1-3 | | Descriptor: | Outer membrane protein assembly factor BamA | | Authors: | Noinaj, N, Lukacik, P, Chang, H, Easley, N, Buchanan, S.K. | | Deposit date: | 2013-04-10 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structural insight into the biogenesis of beta-barrel membrane proteins.
Nature, 501, 2013
|
|
2MBF
 
 | |
4K3B
 
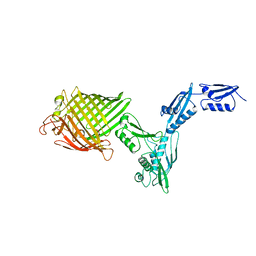 | | The crystal structure of BamA from Neisseria gonorrhoeae | | Descriptor: | Outer membrane protein assembly factor BamA | | Authors: | Noinaj, N, Lukacik, P, Chang, H, Easley, N, Buchanan, S.K. | | Deposit date: | 2013-04-10 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into the biogenesis of beta-barrel membrane proteins.
Nature, 501, 2013
|
|
2M3D
 
 | |
4K5R
 
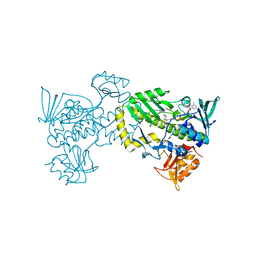 | | The 2.0 angstrom crystal structure of MTMOIV, a baeyer-villiger monooxygenase from the mithramycin biosynthetic pathway in streptomyces argillaceus. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Oxygenase | | Authors: | Noinaj, N, Bosserman, M.A, Rohr, J, Buchanan, S.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Insight into Substrate Recognition and Catalysis of Baeyer-Villiger Monooxygenase MtmOIV, the Key Frame-Modifying Enzyme in the Biosynthesis of Anticancer Agent Mithramycin.
Acs Chem.Biol., 8, 2013
|
|
2MVB
 
 | |
2MQB
 
 | |
