3LQ1
 
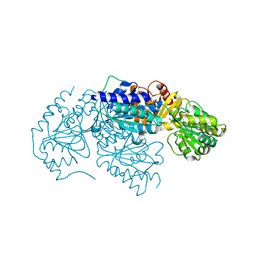 | | Crystal structure of 2-succinyl-6-hydroxy-2,4-cyclohexadiene 1-carboxylic acid synthase/2-oxoglutarate decarboxylase FROM Listeria monocytogenes str. 4b F2365 | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Hu, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Sephchc Synthase from Listeria Monocytogenes
To be Published
|
|
3LKB
 
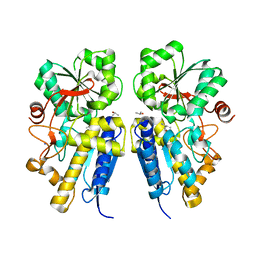 | |
1AJ4
 
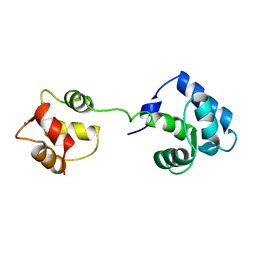 | | STRUCTURE OF CALCIUM-SATURATED CARDIAC TROPONIN C, NMR, 1 STRUCTURE | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Sia, S.K, Li, M.X, Spyracopoulos, L, Gagne, S.M, Liu, W, Putkey, J.A, Sykes, B.D. | | Deposit date: | 1997-05-14 | | Release date: | 1998-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain.
J.Biol.Chem., 272, 1997
|
|
3L6U
 
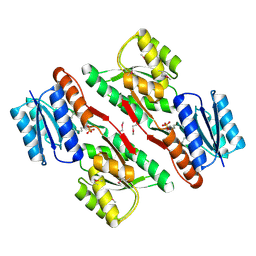 | | Crystal structure of abc-type sugar transport system, Periplasmic component from exiguobacterium sibiricum | | Descriptor: | ABC-TYPE SUGAR TRANSPORT SYSTEM PERIPLASMIC COMPONENT, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-25 | | Release date: | 2010-01-12 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of abc-type sugar transport system,
Periplasmic component from exiguobacterium sibiricum
To be Published
|
|
3LF0
 
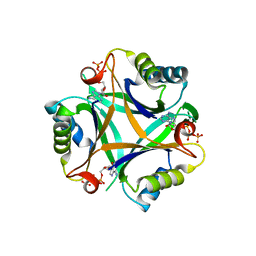 | | Crystal structure of the ATP bound Mycobacterium tuberculosis nitrogen regulatory PII protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Nitrogen regulatory protein P-II | | Authors: | Shetty, N.D, Palaninathan, S.K, Reddy, M.C.M, Owen, J.L, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-01-15 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the apo and ATP bound Mycobacterium tuberculosis nitrogen regulatory PII protein.
Protein Sci., 19, 2010
|
|
3PU5
 
 | |
2HUG
 
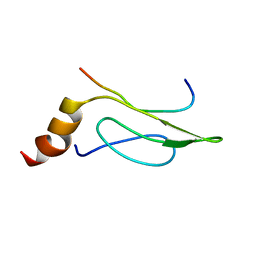 | | 3D Solution Structure of the Chromo-2 Domain of cpSRP43 complexed with cpSRP54 peptide | | Descriptor: | Signal recognition particle 43 kDa protein, chloroplast, Signal recognition particle 54 kDa protein | | Authors: | Kathir, K.M, Vaithiyalingam, S, Henry, R, Thallapuranam, S.K.K. | | Deposit date: | 2006-07-26 | | Release date: | 2007-09-18 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Assembly of chloroplast signal recognition particle involves structural rearrangement in cpSRP43.
J.Mol.Biol., 381, 2008
|
|
3LMD
 
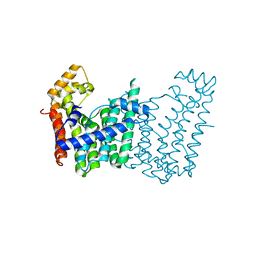 | | Crystal structure of geranylgeranyl pyrophosphate synthase from corynebacterium glutamicum atcc 13032 | | Descriptor: | Geranylgeranyl pyrophosphate synthase | | Authors: | Patskovsky, Y, Ho, M, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Corynebacterium Glutamicum
To be Published
|
|
3LPM
 
 | | Crystal structure of putative methyltransferase small domain protein from Listeria monocytogenes | | Descriptor: | Putative methyltransferase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-05 | | Release date: | 2010-02-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of putative methyltransferase small domain protein from Listeria monocytogenes
To be Published
|
|
3QCY
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 4-[2-Amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-N-phenyl-2-morpholinecarboxamide | | Descriptor: | (2S)-4-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-N-phenylmorpholine-2-carboxamide, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QCX
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-{2-Amino-6-[(3R)-3-methyl-4-morpholinyl]-4-pyrimidinyl}-1H-indazol-3-amine | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-{2-amino-6-[(3R)-3-methylmorpholin-4-yl]pyrimidin-4-yl}-2H-indazol-3-amine, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3LJI
 
 | | CRYSTAL STRUCTURE OF putative geranyltranstransferase from Pseudomonas fluorescens Pf-5 | | Descriptor: | Geranyltranstransferase | | Authors: | Malashkevich, V.N, Toro, R, Patskovsky, Y, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-26 | | Release date: | 2010-02-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | CRYSTAL STRUCTURE OF putative geranyltranstransferase from Pseudomonas fluorescens Pf-5
To be Published
|
|
3QNM
 
 | | Haloalkane Dehalogenase Family Member from Bacteroides thetaiotaomicron of Unknown Function | | Descriptor: | CHLORIDE ION, Haloacid dehalogenase-like hydrolase, MAGNESIUM ION | | Authors: | Matthew, M.W, Ramagopal, U.A, Toro, R, Dickey, M, Sauder, J.M, Poulter, C.D, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-02-08 | | Release date: | 2011-03-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Haloalkane Dehalogenase Family Member from Bacteroides thetaiotaomicron of Unknown Function
To be Published
|
|
3LNV
 
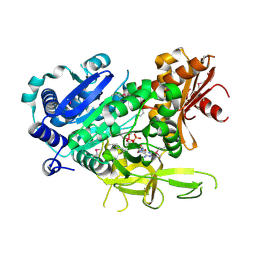 | | The crystal structure of fatty acyl-adenylate ligase from L. pneumophila in complex with acyl adenylate and pyrophosphate | | Descriptor: | 5'-O-[(S)-(dodecanoyloxy)(hydroxy)phosphoryl]adenosine, PYROPHOSPHATE 2-, Saframycin Mx1 synthetase B | | Authors: | Zhang, Z, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-03 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Studies of Fatty Acyl Adenylate Ligases from E. coli and L. pneumophila.
J.Mol.Biol., 406, 2011
|
|
3LOP
 
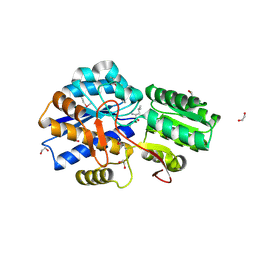 | | Crystal structure of substrate-binding periplasmic protein (Pbp) from Ralstonia solanacearum | | Descriptor: | 1,2-ETHANEDIOL, LEUCINE, MAGNESIUM ION, ... | | Authors: | Palani, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-04 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of substrate-binding periplasmic protein (Pbp) from Ralstonia solanacearum
To be Published
|
|
3LKE
 
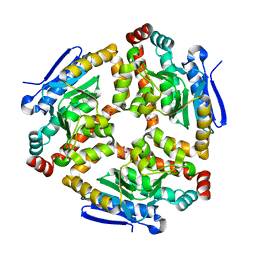 | | Crystal structure of enoyl-CoA hydratase from Bacillus halodurans | | Descriptor: | Enoyl-CoA hydratase, GLYCEROL | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of enoyl-CoA hydratase from Bacillus halodurans
To be Published
|
|
3L60
 
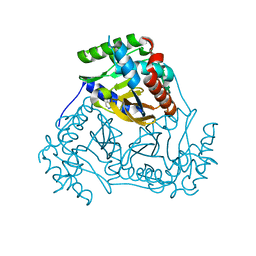 | | Crystal structure of branched-chain alpha-keto acid dehydrogenase subunit e2 from mycobacterium tuberculosis | | Descriptor: | BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE, UNKNOWN LIGAND | | Authors: | Zencheck, W.D, Bonanno, J.B, Patskovsky, Y, Toro, R, Freeman, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Branched-Chain Alpha-Keto Acid Dehydrogenase Subunit E2 from Mycobacterium Tuberculosis
To be Published
|
|
3Q7O
 
 | |
3L8D
 
 | | Crystal structure of methyltransferase from Bacillus Thuringiensis | | Descriptor: | Methyltransferase, POTASSIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of methyltransferase from Bacillus Thuringiensis
To be Published
|
|
3L8K
 
 | | Crystal structure of a dihydrolipoyl dehydrogenase from Sulfolobus solfataricus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dihydrolipoyl dehydrogenase, PHOSPHATE ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a dihydrolipoyl dehydrogenase from Sulfolobus solfataricus
To be Published
|
|
3Q34
 
 | |
3Q4D
 
 | | Crystal structure of dipeptide epimerase from Cytophaga hutchinsonii complexed with Mg and dipeptide D-Ala-L-Ala | | Descriptor: | ALANINE, D-ALANINE, MAGNESIUM ION, ... | | Authors: | Lukk, T, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2010-12-23 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8SE9
 
 | | Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex (Form 2) | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | Authors: | Afsar, M, Jia, L, Ruben, E.A, Olsen, S.K. | | Deposit date: | 2023-04-08 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer.
Nat Commun, 14, 2023
|
|
3LTO
 
 | |
3POP
 
 | | The crystal structure of GilR, an oxidoreductase that catalyzes the terminal step of gilvocarcin biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GilR oxidase | | Authors: | Noinaj, N, Bosserman, M.A, Schickli, M.A, Kharel, M.K, Rohr, J, Buchanan, S.K. | | Deposit date: | 2010-11-23 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | The Crystal Structure and Mechanism of an Unusual Oxidoreductase, GilR, Involved in Gilvocarcin V Biosynthesis.
J.Biol.Chem., 286, 2011
|
|
