7UZF
 
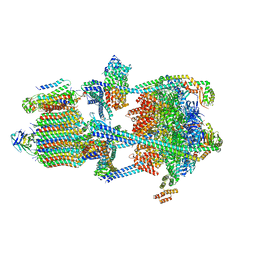 | | Rat Kidney V-ATPase with SidK, State 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H+-transporting V1 subunit A, ATPase H+-transporting V1 subunit D, ... | | Authors: | Rubinstein, J.L, Abbas, Y.M. | | Deposit date: | 2022-05-09 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of V-ATPase with NCOA7B
To Be Published
|
|
7UZI
 
 | |
7UZH
 
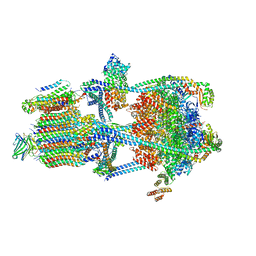 | | Rat Kidney V-ATPase with SidK, State 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H+-transporting V1 subunit A, ATPase H+-transporting V1 subunit D, ... | | Authors: | Rubinstein, J.L, Abbas, Y.M. | | Deposit date: | 2022-05-09 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of V-ATPase with NCOA7B
To Be Published
|
|
7UZG
 
 | |
7Q21
 
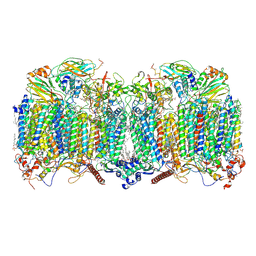 | | III2-IV2 respiratory supercomplex from Corynebacterium glutamicum | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Kovalova, T, Moe, A, Krol, S, Yanofsky, D.J, Bott, M, Sjostrand, D, Rubinstein, J.L, Hogbom, M, Brzezinski, P. | | Deposit date: | 2021-10-22 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The respiratory supercomplex from C. glutamicum.
Structure, 30, 2022
|
|
9MQZ
 
 | |
9MQY
 
 | |
9DM1
 
 | | Mycobacterial supercomplex malate:quinone oxidoreductase assembly | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Di Trani, J.M, Rubinstein, J.L. | | Deposit date: | 2024-09-11 | | Release date: | 2024-10-23 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM of native membranes reveals an intimate connection between the Krebs cycle and aerobic respiration in mycobacteria.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9BRD
 
 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3 | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coupland, C.E, Rubinstein, J.L. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
9BRB
 
 | |
9BRC
 
 | |
9B8O
 
 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3, Vo | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coupland, C.E, Rubinstein, J.L. | | Deposit date: | 2024-03-31 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
9B8Q
 
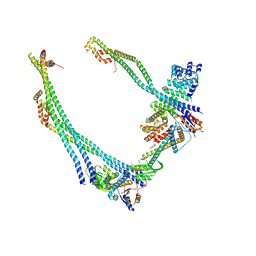 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3, peripheral stalks | | Descriptor: | V-type proton ATPase 116 kDa subunit a isoform 1, V-type proton ATPase subunit C 1, V-type proton ATPase subunit E 1, ... | | Authors: | Coupland, C.E, Rubinstein, J.L. | | Deposit date: | 2024-03-31 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
9B8P
 
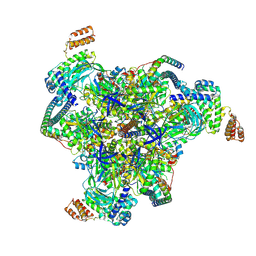 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3, V1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H+-transporting V1 subunit D, H(+)-transporting two-sector ATPase, ... | | Authors: | Coupland, E.M, Rubinstein, J.L. | | Deposit date: | 2024-03-31 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
9COP
 
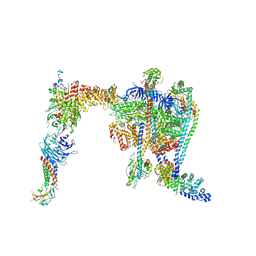 | | Yeast RAVE bound to V-ATPase V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Regulator of V-ATPase in vacuolar membrane protein 1, ... | | Authors: | Wang, H, Rubinstein, J.L. | | Deposit date: | 2024-07-17 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of yeast RAVE bound to a partial V 1 complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6HWH
 
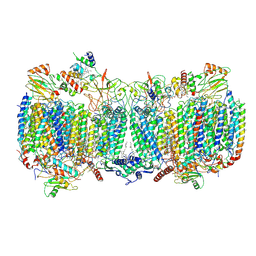 | | Structure of a functional obligate respiratory supercomplex from Mycobacterium smegmatis | | Descriptor: | CARDIOLIPIN, COPPER (II) ION, Co-purified unknown peptide built as polyALA, ... | | Authors: | Wiseman, B, Nitharwal, R.G, Fedotovskaya, O, Schafer, J, Guo, H, Kuang, Q, Benlekbir, S, Sjostrand, D, Adelroth, P, Rubinstein, J.L, Brzezinski, P, Hogbom, M. | | Deposit date: | 2018-10-12 | | Release date: | 2018-11-07 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a functional obligate complex III2IV2respiratory supercomplex from Mycobacterium smegmatis.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6O7V
 
 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 1 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6O7W
 
 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 2 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7MD2
 
 | | The F1 region of ammocidin-bound Saccharomyces cerevisiae ATP synthase | | Descriptor: | (3~{E},5~{Z},7~{E},9~{R},10~{S},11~{E},13~{E},15~{E},17~{R},18~{S},20~{S})-20-[(1~{R})-1-[(2~{S},3~{R},4~{R},5~{S},6~{R})-5-[(2~{S},4~{S},5~{S},6~{R})-5-[(2~{S},4~{R},5~{R},6~{R})-4,6-dimethyl-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-4-oxidanyl-oxan-2-yl]oxy-3-methoxy-6-(3-methoxypropyl)-5-methyl-2,4-bis(oxidanyl)oxan-2-yl]ethyl]-5,18-dimethoxy-3,7,9,11,13,15-hexamethyl-10-[(2~{R},3~{S},4~{R},5~{R},6~{S})-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-17-oxidanyl-1-oxacycloicosa-3,5,7,11,13,15-hexaen-2-one, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2021-04-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A Family of Glycosylated Macrolides Selectively Target Energetic Vulnerabilities in Leukemia
Biorxiv, 2021
|
|
7MD3
 
 | | The F1 region of apoptolidin-bound Saccharomyces cerevisiae ATP synthase | | Descriptor: | (3~{E},5~{E},7~{E},9~{R},10~{R},11~{E},13~{E},17~{S},18~{S},20~{S})-18-methoxy-20-[(~{R})-[(2~{R},3~{R},4~{S},5~{R},6~{R})-6-[(2~{R})-3-methoxy-2-[(2~{R},4~{S},5~{S},6~{S})-5-[(2~{S},4~{R},5~{R},6~{R})-4-methoxy-6-methyl-5-oxidanyl-oxan-2-yl]oxy-4,6-dimethyl-4-oxidanyl-oxan-2-yl]oxy-propyl]-3,5-dimethyl-2,4-bis(oxidanyl)oxan-2-yl]-oxidanyl-methyl]-10-[(2~{R},3~{S},4~{S},5~{R},6~{S})-5-methoxy-6-methyl-3,4-bis(oxidanyl)oxan-2-yl]oxy-3,5,7,9,13-pentamethyl-17-oxidanyl-1-oxacycloicosa-3,5,7,11,13-pentaen-2-one, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2021-04-03 | | Release date: | 2021-08-11 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A Family of Glycosylated Macrolides Selectively Target Energetic Vulnerabilities in Leukemia
Biorxiv, 2021
|
|
7MPE
 
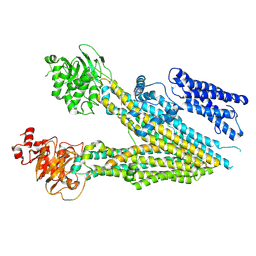 | |
5GAS
 
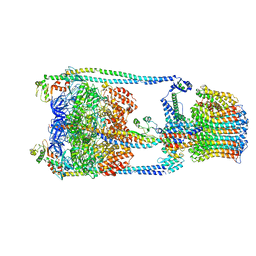 | | Thermus thermophilus V/A-ATPase, conformation 2 | | Descriptor: | Archaeal/vacuolar-type H+-ATPase subunit I, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Schep, D.G, Zhao, J, Rubinstein, J.L. | | Deposit date: | 2016-02-05 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5G4G
 
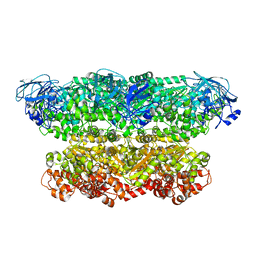 | | Structure of the ATPgS-bound VAT complex | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Huang, R, Ripstein, Z.A, Augustyniak, R, Lazniewski, M, Ginalski, K, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Unfolding the Mechanism of the Aaa+ Unfoldase Vat by a Combined Cryo-Em, Solution NMR Study.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5G4F
 
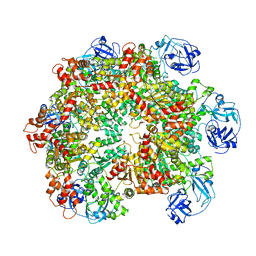 | | Structure of the ADP-bound VAT complex | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Huang, R, Ripstein, Z.A, Augustyniak, R, Lazniewski, M, Ginalski, K, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Unfolding the Mechanism of the Aaa+ Unfoldase Vat by a Combined Cryo-Em, Solution NMR Study.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GAR
 
 | | Thermus thermophilus V/A-ATPase, conformation 1 | | Descriptor: | Archaeal/vacuolar-type H+-ATPase subunit I, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Schep, D.G, Zhao, J, Rubinstein, J.L. | | Deposit date: | 2016-02-05 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
