6TOP
 
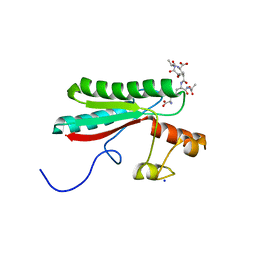 | | Structure of the PorE C-terminal domain, a protein of T9SS from Porphyromonas gingivalis | | Descriptor: | GLCNAC(BETA1-4)-MURNAC(1,6-ANHYDRO)-L-ALA-GAMMA-D-GLU-MESO-A2PM-D-ALA, OmpA family protein, SODIUM ION | | Authors: | Trinh, T.N, Cambillau, C, Roussel, A, Leone, P. | | Deposit date: | 2019-12-11 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of Type IX secretion system PorE C-terminal domain from Porphyromonas gingivalis in complex with a peptidoglycan fragment.
Sci Rep, 10, 2020
|
|
2CYG
 
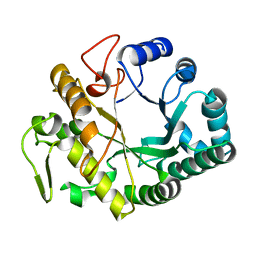 | | Crystal structure at 1.45- resolution of the major allergen endo-beta-1,3-glucanase of banana as a molecular basis for the latex-fruit syndrome | | Descriptor: | beta-1, 3-glucananse | | Authors: | Receveur-Brechot, V, Czjzek, M, Barre, A, Roussel, A, Peumans, W.J, Van Damme, E.J.M, Rouge, P. | | Deposit date: | 2005-07-07 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure at 1.45-A resolution of the major allergen endo-beta-1,3-glucanase of banana as a molecular basis for the latex-fruit syndrome
Proteins, 63, 2006
|
|
4MKQ
 
 | |
4MKO
 
 | |
4MJT
 
 | |
4MNS
 
 | | Crystal structure of the major pollen allergen Bet v 1-A in complex with P303 | | Descriptor: | (3R,4R,5aR,11aR)-3-methyl-6,11-dioxo-2,3,4,5,5a,6,11,11a-octahydrothiepino[3,2-g]isoquinoline-4-carboxylic acid, Major pollen allergen Bet v 1-A | | Authors: | Leone, P, Roussel, A. | | Deposit date: | 2013-09-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Development and evaluation of a sublingual tablet based on recombinant Bet v 1 in birch pollen-allergic patients.
Allergy, 70, 2015
|
|
1WO9
 
 | | Selective inhibition of trypsins by insect peptides: role of P6-P10 loop | | Descriptor: | trypsin inhibitor | | Authors: | Kellenberger, C, Ferrat, G, Leone, P, Darbon, H, Roussel, A. | | Deposit date: | 2004-08-12 | | Release date: | 2004-09-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Selective inhibition of trypsins by insect peptides: role of P6-P10 loop
Biochemistry, 42, 2003
|
|
5LMJ
 
 | | Llama nanobody PorM_19 | | Descriptor: | PHOSPHATE ION, nanobody | | Authors: | Leone, P, Cambillau, C, Roussel, A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Camelid nanobodies used as crystallization chaperones for different constructs of PorM, a component of the type IX secretion system from Porphyromonas gingivalis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5LMW
 
 | | Llama nanobody PorM_02 | | Descriptor: | GLYCEROL, Nanobody | | Authors: | Roche, J, Gaubert, A, Leone, P, Roussel, A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Camelid nanobodies used as crystallization chaperones for different constructs of PorM, a component of the type IX secretion system from Porphyromonas gingivalis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5MP2
 
 | | XcpQN012 in complex with VHH04 | | Descriptor: | Camelid nanobody VHH04, Type II secretion system protein D | | Authors: | Trinh, T.T.N, Roussel, A, Douzi, B, Voulhoux, R. | | Deposit date: | 2016-12-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Unraveling the Self-Assembly of the Pseudomonas aeruginosa XcpQ Secretin Periplasmic Domain Provides New Molecular Insights into Type II Secretion System Secreton Architecture and Dynamics.
MBio, 8, 2017
|
|
5LZ0
 
 | | Llama nanobody PorM_01 | | Descriptor: | PorM nanobody | | Authors: | Duhoo, Y, Leone, P, Roussel, A. | | Deposit date: | 2016-09-29 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Camelid nanobodies used as crystallization chaperones for different constructs of PorM, a component of the type IX secretion system from Porphyromonas gingivalis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5NGI
 
 | | Structure of XcpQN012 | | Descriptor: | Type II secretion system protein D | | Authors: | Cambillau, C, Roussel, A, Trinh, T.N. | | Deposit date: | 2017-03-17 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Unraveling the Self-Assembly of the Pseudomonas aeruginosa XcpQ Secretin Periplasmic Domain Provides New Molecular Insights into Type II Secretion System Secreton Architecture and Dynamics.
MBio, 8, 2017
|
|
3ZIZ
 
 | | Crystal structure of Podospora anserina GH5 beta-(1,4)-mannanase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GH5 ENDO-BETA-1,4-MANNANASE, GLYCEROL | | Authors: | Couturier, M, Roussel, A, Rosengren, A, Leone, P, Stalbrand, H, Berrin, J.G. | | Deposit date: | 2013-01-15 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Families 5 and 26 Beta-(1,4)-Mannanases from Podospora Anserina Reveal Differences Upon Manno-Oligosaccharides Catalysis.
J.Biol.Chem., 288, 2013
|
|
3ZM8
 
 | | Crystal structure of Podospora anserina GH26-CBM35 beta-(1,4)- mannanase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GH26 ENDO-BETA-1,4-MANNANASE, ... | | Authors: | Couturier, M, Roussel, A, Rosengren, A, Leone, P, Stalbrand, H, Berrin, J.G. | | Deposit date: | 2013-02-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Families 5 and 26 Beta-(1,4)-Mannanases from Podospora Anserina Reveal Differences Upon Manno-Oligosaccharides Catalysis.
J.Biol.Chem., 288, 2013
|
|
1KX9
 
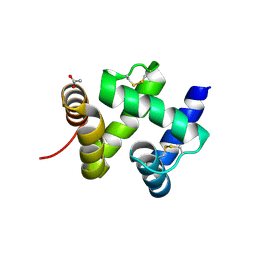 | | ANTENNAL CHEMOSENSORY PROTEIN A6 FROM THE MOTH MAMESTRA BRASSICAE | | Descriptor: | ACETATE ION, CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structure and ligand binding study of a moth chemosensory protein
J.Biol.Chem., 277, 2002
|
|
1KX8
 
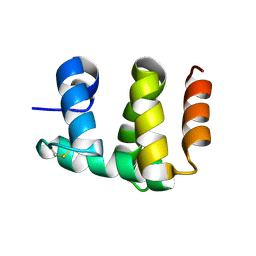 | | Antennal Chemosensory Protein A6 from Mamestra brassicae, tetragonal form | | Descriptor: | CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray Structure and Ligand Binding Study of a Chemosensory Protein
J.Biol.Chem., 277, 2002
|
|
2CF4
 
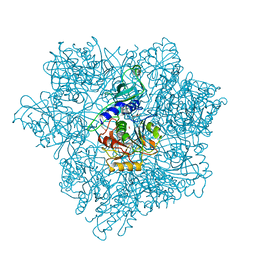 | | Pyrococcus horikoshii TET1 peptidase can assemble into a tetrahedron or a large octahedral shell | | Descriptor: | COBALT (II) ION, PROTEIN PH0519 | | Authors: | Vellieux, F.M.D, Schoehn, G, Dura, M.A, Roussel, A, Franzetti, B. | | Deposit date: | 2006-02-15 | | Release date: | 2006-09-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | An Archaeal Peptidase Assembles Into Two Different Quaternary Structures: A Tetrahedron and a Giant Octahedron.
J.Biol.Chem., 281, 2006
|
|
2VU8
 
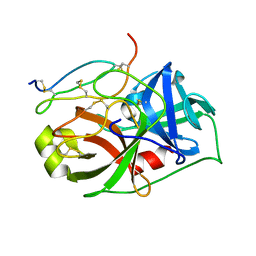 | |
3ZZO
 
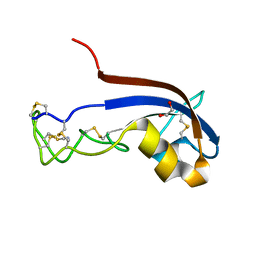 | |
3ZZR
 
 | | Crystal structure of the CG11501 protein in P21212 spacegroup | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Coste, F, Roussel, A. | | Deposit date: | 2011-09-02 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Diedel, a Marker of the Immune Response of Drosophila Melanogaster.
Plos One, 7, 2012
|
|
1I9W
 
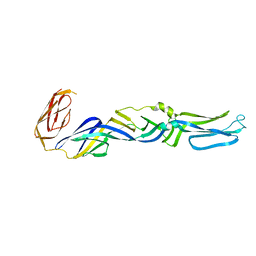 | | CRYSTAL STRUCTURE OF THE FUSION GLYCOPROTEIN E1 FROM SEMLIKI FOREST VIRUS | | Descriptor: | FUSION PROTEIN E1 | | Authors: | Lescar, J, Roussel, A, Wien, M.W, Navaza, J, Fuller, S.D, Wengler, G, Wengler, G, Rey, F.A. | | Deposit date: | 2001-03-21 | | Release date: | 2002-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH.
Cell(Cambridge,Mass.), 105, 2001
|
|
1LK5
 
 | | Structure of the D-Ribose-5-Phosphate Isomerase from Pyrococcus horikoshii | | Descriptor: | CHLORIDE ION, D-Ribose-5-Phosphate Isomerase, SODIUM ION | | Authors: | Ishikawa, K, Matsui, I, Payan, F, Cambillau, C, Ishida, H, Kawarabayasi, Y, Kikuchi, H, Roussel, A. | | Deposit date: | 2002-04-24 | | Release date: | 2002-07-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A hyperthermostable D-ribose-5-phosphate isomerase from Pyrococcus horikoshii characterization and three-dimensional structure.
Structure, 10, 2002
|
|
1LK7
 
 | | Structure of D-Ribose-5-Phosphate Isomerase from in complex with phospho-erythronic acid | | Descriptor: | CHLORIDE ION, D-4-PHOSPHOERYTHRONIC ACID, D-Ribose-5-Phosphate Isomerase, ... | | Authors: | Ishikawa, K, Matsui, I, Payan, F, Cambillau, C, Ishida, H, Kawarabayasi, Y, Kikuchi, H, Roussel, A. | | Deposit date: | 2002-04-24 | | Release date: | 2002-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Hyperthermostable D-Ribose-5-Phosphate Isomerase from Pyrococcus horikoshii Characterization and Three-Dimensional Structure
STRUCTURE, 10, 2002
|
|
