5FWO
 
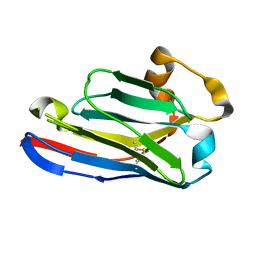 | | Llama nanobody PorM_130 | | Descriptor: | NANOBODY | | Authors: | Leone, P, Roussel, A. | | Deposit date: | 2016-02-18 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Camelid nanobodies used as crystallization chaperones for different constructs of PorM, a component of the type IX secretion system from Porphyromonas gingivalis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6EY0
 
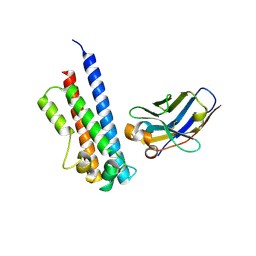 | | N-terminal part (residues 30-212) of PorM with the llama nanobody nb01 | | Descriptor: | T9SS component cytoplasmic membrane protein PorM, llama nanobody nb01 | | Authors: | Leone, P, Roche, J, Cambillau, C, Roussel, A. | | Deposit date: | 2017-11-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space.
Nat Commun, 9, 2018
|
|
6EY6
 
 | |
3IE4
 
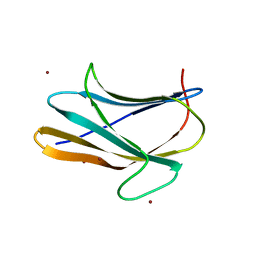 | | b-glucan binding domain of Drosophila GNBP3 defines a novel family of pattern recognition receptor | | Descriptor: | 1,2-ETHANEDIOL, Gram-Negative Binding Protein 3, ZINC ION | | Authors: | Mishima, Y, Coste, F, Kellenberger, C, Roussel, A. | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The N-terminal domain of drosophila gram-negative binding protein 3 (GNBP3) defines a novel family of fungal pattern recognition receptors
To be Published
|
|
6EY5
 
 | | C-terminal part (residues 224-515) of PorM | | Descriptor: | T9SS component cytoplasmic membrane protein PorM, ZINC ION | | Authors: | Leone, P, Cambillau, C, Roussel, A. | | Deposit date: | 2017-11-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space.
Nat Commun, 9, 2018
|
|
5EFV
 
 | | The host-recognition device of Staphylococcus aureus phage Phi11 | | Descriptor: | FE (III) ION, Phi ETA orf 56-like protein | | Authors: | Koc, C, Kuhner, P, Xia, G, Spinelli, S, Roussel, A, Cambillau, C, Stehle, T. | | Deposit date: | 2015-10-26 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the host-recognition device of Staphylococcus aureus phage 11.
Sci Rep, 6, 2016
|
|
1RER
 
 | | Crystal structure of the homotrimer of fusion glycoprotein E1 from Semliki Forest Virus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gibbons, D.L, Vaney, M.C, Roussel, A, Vigouroux, A, Reilly, B, Kielian, M, Rey, F.A. | | Deposit date: | 2003-11-07 | | Release date: | 2004-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus.
Nature, 427, 2004
|
|
4GI3
 
 | |
1ZT7
 
 | | crystal structure of class I MHC H-2Kk in complex with a nonapeptide | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-K alpha chain, ... | | Authors: | Kellenberger, C, Roussel, A, Malissen, B. | | Deposit date: | 2005-05-26 | | Release date: | 2005-10-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The H-2Kk MHC peptide-binding groove anchors the backbone of an octameric antigenic peptide in an unprecedented mode.
J Immunol., 175, 2005
|
|
1ZT1
 
 | | crystal structure of class I MHC H-2Kk in complex with an octapeptide | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, H-2 class I histocompatibility antigen, ... | | Authors: | Kellenberger, C, Roussel, A, Malissen, B. | | Deposit date: | 2005-05-26 | | Release date: | 2005-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The H-2Kk MHC peptide-binding groove anchors the backbone of an octameric antigenic peptide in an unprecedented mode.
J Immunol., 175, 2005
|
|
6ZPP
 
 | |
8BOZ
 
 | |
1Z3Q
 
 | | Resolution of the structure of the allergenic and antifungal banana fruit thaumatin-like protein at 1.7A | | Descriptor: | 1,2-ETHANEDIOL, Thaumatin-like Protein | | Authors: | Leone, P, Menu-Bouaouiche, L, Peumans, W.J, Barre, A, Payan, F, Roussel, A, Van Damme, E.J.M, Rouge, P. | | Deposit date: | 2005-03-14 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Resolution of the structure of the allergenic and antifungal banana fruit thaumatin-like protein at 1.7-A
Biochimie, 88, 2006
|
|
6EY4
 
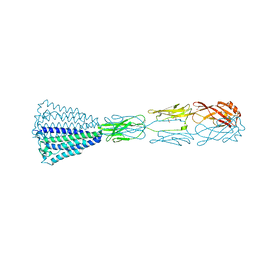 | | Periplasmic domain (residues 36-513) of GldM | | Descriptor: | 1,2-ETHANEDIOL, GldM | | Authors: | Leone, P, Roche, J, Cambillau, C, Roussel, A. | | Deposit date: | 2017-11-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space.
Nat Commun, 9, 2018
|
|
4QGY
 
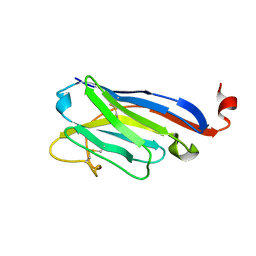 | | Camelid (llama) nanobody n25 (VHH) against type 6 secretion system TssM protein | | Descriptor: | nanobody n25, VH domain | | Authors: | Nguyen, V.S, Desmyter, A, Le, T.T.H, Durand, E, Kellenberger, C, Douzi, B, Spinelli, S, Cascales, E, Cambillau, C, Roussel, A. | | Deposit date: | 2014-05-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Inhibition of Type VI Secretion by an Anti-TssM Llama Nanobody.
Plos One, 10, 2015
|
|
4QLR
 
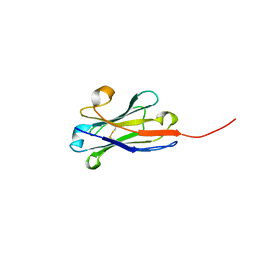 | | Llama nanobody n02 raised against EAEC T6SS TssM | | Descriptor: | Llama nanobody n02 VH domain | | Authors: | Nguyen, V.S, Desmyter, A, Le, T.T.H, Durand, E, Kellenberger, C, Douzi, B, Spinelli, S, Cascales, E, Cambillau, C, Roussel, A. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Type VI Secretion by an Anti-TssM Llama Nanobody.
Plos One, 10, 2015
|
|
2XXL
 
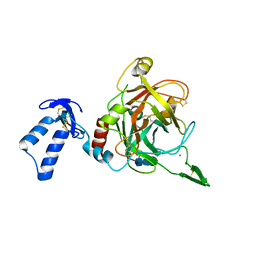 | | Crystal structure of drosophila Grass clip serine protease of Toll pathway | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GRAM-POSITIVE SPECIFIC SERINE PROTEASE, ... | | Authors: | Kellenberger, C, Leone, P, Coquet, L, Jouenne, T, Reichhart, J.M, Roussel, A. | | Deposit date: | 2010-11-10 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Analysis of Grass Clip Serine Protease Involved in Drosophila Toll Pathway Activation.
J.Biol.Chem., 286, 2011
|
|
2XZ8
 
 | | CRYSTAL STRUCTURE OF THE LFW ECTODOMAIN OF THE PEPTIDOGLYCAN RECOGNITION PROTEIN LF | | Descriptor: | DI(HYDROXYETHYL)ETHER, PEPTIDOGLYCAN-RECOGNITION PROTEIN LF, SODIUM ION, ... | | Authors: | Basbous, N, Coste, F, Leone, P, Vincentelli, R, Royet, J, Kellenberger, C, Roussel, A. | | Deposit date: | 2010-11-24 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Drosophila Peptidoglycan-Recognition Protein Lf Interacts with Peptidoglycan-Recognition Protein Lc to Downregulate the Imd Pathway.
Embo Rep., 12, 2011
|
|
2XZ4
 
 | | Crystal structure of the LFZ ectodomain of the peptidoglycan recognition protein LF | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, COPPER (II) ION, ... | | Authors: | Basbous, N, Coste, F, Leone, P, Vincentelli, R, Royet, J, Kellenberger, C, Roussel, A. | | Deposit date: | 2010-11-23 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The Drosophila Peptidoglycan-Recognition Protein Lf Interacts with Peptidoglycan-Recognition Protein Lc to Downregulate the Imd Pathway.
Embo Rep., 12, 2011
|
|
6GZP
 
 | | Llama nanobody PorM_02 structure determined at room temperature by in-situ diffraction in ChipX microfluidic device | | Descriptor: | Nanobody | | Authors: | Roche, J, Gaubert, A, Desmyter, A, De Wijn, R, Sauter, C, Roussel, A. | | Deposit date: | 2018-07-04 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
1CSJ
 
 | | CRYSTAL STRUCTURE OF THE RNA-DEPENDENT RNA POLYMERASE OF HEPATITIS C VIRUS | | Descriptor: | HEPATITIS C VIRUS RNA POLYMERASE (NS5B) | | Authors: | Bressanelli, S, Tomei, L, Roussel, A, Incitti, I, Vitale, R.L, Mathieu, M, De Francesco, R, Rey, F.A. | | Deposit date: | 1999-08-18 | | Release date: | 1999-11-08 | | Last modified: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2CLZ
 
 | | Mhc Class I Natural Mutant H-2Kbm8 Heavy Chain Complexed With beta-2 Microglobulin and pBM1 peptide | | Descriptor: | BETA-2 MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, K-B ALPHA CHAIN, ... | | Authors: | Mazza, C, Auphan-Anezin, N, Guimezanes, A, Barrett-Wilt, G.A, Montero-Julian, F, Roussel, A, Hunt, D.F, Schmitt-Verhulst, A.M, Malissen, B. | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct Orientation of the Alloreactive Monoclonal Cd8 T Cell Activation Program by Three Different Peptide/Mhc Complexes.
Eur.J.Immunol., 36, 2006
|
|
2CLV
 
 | | MHC Class I Natural Mutant H-2Kbm8 Heavy Chain Complexed With beta-2 Microglobulin and pBM8 peptide | | Descriptor: | BETA-2 MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, K-B ALPHA CHAIN, ... | | Authors: | Mazza, C, Auphan-Anezin, N, Guimezanes, A, Barrett-Wilt, G.A, Montero-Julian, F, Roussel, A, Hunt, D.F, Schmitt-Verhulst, A.M, Malissen, B. | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct orientation of the alloreactive monoclonal CD8 T cell activation program by three different peptide/MHC complexes.
Eur. J. Immunol., 36, 2006
|
|
1TE1
 
 | | Crystal structure of family 11 xylanase in complex with inhibitor (XIP-I) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, endo-1,4-xylanase, ... | | Authors: | Payan, F, Leone, P, Furniss, C, Tahir, T, Durand, A, Porciero, S, Manzanares, P, Williamson, G, Gilbert, H.J, Juge, N, Roussel, A. | | Deposit date: | 2004-05-24 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Dual Nature of the Wheat Xylanase Protein Inhibitor XIP-I: STRUCTURAL BASIS FOR THE INHIBITION OF FAMILY 10 AND FAMILY 11 XYLANASES.
J.Biol.Chem., 279, 2004
|
|
1TA3
 
 | | Crystal Structure of xylanase (GH10) in complex with inhibitor (XIP) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Endo-1,4-beta-xylanase, ... | | Authors: | Payan, F, Leone, P, Furniss, C, Tahir, T, Durand, A, Porciero, S, Manzanares, P, Williamson, G, Gilbert, H.J, Juge, N, Roussel, A. | | Deposit date: | 2004-05-19 | | Release date: | 2004-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Dual Nature of the Wheat Xylanase Protein Inhibitor XIP-I: STRUCTURAL BASIS FOR THE INHIBITION OF FAMILY 10 AND FAMILY 11 XYLANASES.
J.Biol.Chem., 279, 2004
|
|
