3ZNU
 
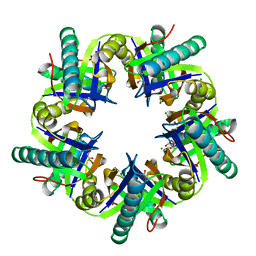 | | Crystal structure of ClcF in crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION, ... | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
3ZNJ
 
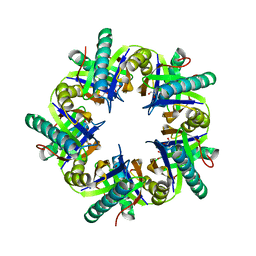 | | Crystal structure of unliganded ClcF from R.opacus 1CP in crystal form 1. | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
6FHV
 
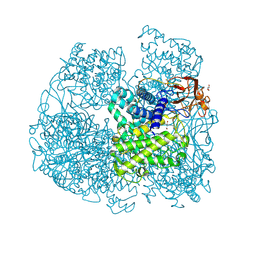 | | Crystal structure of Penicillium oxalicum Glucoamylase | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FHW
 
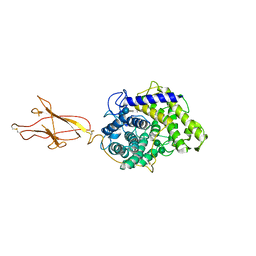 | | Structure of Hormoconis resinae Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5L7V
 
 | | Crystal Structure of BvGH123 with bond transition state analog Galthiazoline. | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, glycoside hydrolase | | Authors: | Roth, C, Petricevic, M, John, A, Goddard-Borger, E.D, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic insights into a Bacteroides vulgatus retaining N-acetyl-beta-galactosaminidase that uses neighbouring group participation.
Chem. Commun. (Camb.), 52, 2016
|
|
5L7U
 
 | | Crystal structure of BvGH123 with bound GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CHLORIDE ION, Glycoside hydrolase | | Authors: | Roth, C, Petricevic, M, John, A, Goddard-Borger, E.D, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic insights into a Bacteroides vulgatus retaining N-acetyl-beta-galactosaminidase that uses neighbouring group participation.
Chem. Commun. (Camb.), 52, 2016
|
|
5L7R
 
 | | Crystal structure of BvGH123 | | Descriptor: | 1,2-ETHANEDIOL, glycoside hydrolase | | Authors: | Roth, C, Petricevic, M, John, A, Goddard-Borger, E.D, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-22 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and mechanistic insights into a Bacteroides vulgatus retaining N-acetyl-beta-galactosaminidase that uses neighbouring group participation.
Chem. Commun. (Camb.), 52, 2016
|
|
6FRV
 
 | | Structure of the catalytic domain of Aspergillus niger Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucoamylase, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-02-16 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5JIW
 
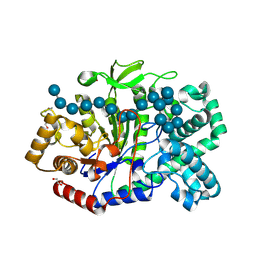 | | Crystal structure of Thermus aquaticus amylomaltase (GH77) in complex with a 34-meric cycloamylose | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase, CARBONATE ION, ... | | Authors: | Roth, C, Bexten, N, Weizenmann, N, Saenger, T, Maier, T, Zimmermann, W, Straeter, N. | | Deposit date: | 2016-04-22 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Amylose recognition and ring-size determination of amylomaltase.
Sci Adv, 3, 2017
|
|
5M7R
 
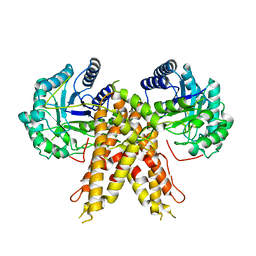 | | Structure of human O-GlcNAc hydrolase | | Descriptor: | Protein O-GlcNAcase | | Authors: | Roth, C, Chan, S, Offen, W.A, Hemsworth, G.R, Willems, L.I, King, D, Varghese, V, Britton, R, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional insight into human O-GlcNAcase.
Nat. Chem. Biol., 13, 2017
|
|
5M7U
 
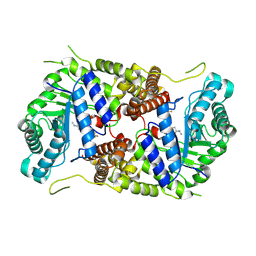 | | Structure of human O-GlcNAc hydrolase with new iminocyclitol type inhibitor | | Descriptor: | 2-[(2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-[3-[3-(trifluoromethyl)phenyl]propyl]pyrrolidin-2-yl]-~{N}-methyl-ethanamide, Protein O-GlcNAcase | | Authors: | Roth, C, Chan, S, Offen, W.A, Hemsworth, G.R, Willems, L.I, King, D, Varghese, V, Britton, R, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional insight into human O-GlcNAcase.
Nat. Chem. Biol., 13, 2017
|
|
5M7T
 
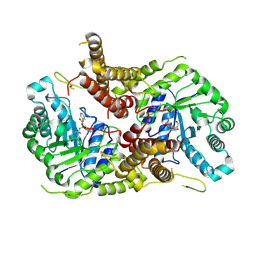 | | Structure of human O-GlcNAc hydrolase with PugNAc type inhibitor | | Descriptor: | (5R,6R,7R,8S)-8-(ACETYLAMINO)-6,7-DIHYDROXY-5-(HYDROXYMETHYL)-N-PHENYL-1,5,6,7,8,8A-HEXAHYDROIMIDAZO[1,2-A]PYRIDINE-2-CARBOXAMIDE, Protein O-GlcNAcase | | Authors: | Roth, C, Chan, S, Offen, W.A, Hemsworth, G.R, Willems, L.I, King, D, Varghese, V, Britton, R, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-29 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional insight into human O-GlcNAcase.
Nat. Chem. Biol., 13, 2017
|
|
5M7S
 
 | | Structure of human O-GlcNAc hydrolase with bound transition state analog ThiametG | | Descriptor: | (3AR,5R,6S,7R,7AR)-2-(ETHYLAMINO)-5-(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, Protein O-GlcNAcase | | Authors: | Roth, C, Chan, S, Offen, W.A, Hemsworth, G.R, Willems, L.I, King, D, Varghese, V, Britton, R, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-29 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insight into human O-GlcNAcase.
Nat. Chem. Biol., 13, 2017
|
|
6SAO
 
 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
6SAV
 
 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, CALCIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
6SAU
 
 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance. | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, SODIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tinaqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
3ZO7
 
 | | Crystal structure of ClcFE27A with substrate | | Descriptor: | (2S)-2-chloranyl-2-[(2R)-5-oxidanylidene-2H-furan-2-yl]ethanoic acid, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.224 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
4CG3
 
 | | Structural and functional studies on a thermostable polyethylene therephtalate degrading hydrolase from Thermobifida fusca | | Descriptor: | CUTINASE, SULFATE ION | | Authors: | Roth, C, Wei, R, Oeser, T, Then, J, Foellner, C, Zimmermann, W, Straeter, N. | | Deposit date: | 2013-11-20 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Functional Studies on a Thermostable Polyethylene Terephthalate Degrading Hydrolase from Thermobifida Fusca.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
4CG1
 
 | | Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca | | Descriptor: | CUTINASE, SULFATE ION | | Authors: | Roth, C, Wei, R, Oeser, T, Then, J, Foellner, C, Zimmermann, W, Straeter, N. | | Deposit date: | 2013-11-20 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Studies on a Thermostable Polyethylene Terephthalate Degrading Hydrolase from Thermobifida Fusca.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
4CG2
 
 | | Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca | | Descriptor: | CUTINASE, SULFATE ION, phenylmethanesulfonic acid | | Authors: | Roth, C, Wei, R, Oeser, T, Then, J, Foellner, C, Zimmermann, W, Straeter, N. | | Deposit date: | 2013-11-20 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.437 Å) | | Cite: | Structural and Functional Studies on a Thermostable Polyethylene Terephthalate Degrading Hydrolase from Thermobifida Fusca.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
2QGI
 
 | |
2NXV
 
 | |
1PN0
 
 | | Phenol hydroxylase from Trichosporon cutaneum | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHENOL, ... | | Authors: | Enroth, C. | | Deposit date: | 2003-06-12 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structure of phenol hydroxylase and correction of sequence errors.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1FO4
 
 | | CRYSTAL STRUCTURE OF XANTHINE DEHYDROGENASE ISOLATED FROM BOVINE MILK | | Descriptor: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Enroth, C, Eger, B.T, Okamoto, K, Nishino, T, Nishino, T, Pai, E.F. | | Deposit date: | 2000-08-24 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FOH
 
 | | PHENOL HYDROXYLASE FROM TRICHOSPORON CUTANEUM | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHENOL, PHENOL HYDROXYLASE | | Authors: | Enroth, C, Neujahr, H, Schneider, G, Lindqvist, Y. | | Deposit date: | 1998-03-26 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of phenol hydroxylase in complex with FAD and phenol provides evidence for a concerted conformational change in the enzyme and its cofactor during catalysis.
Structure, 6, 1998
|
|
