5MWV
 
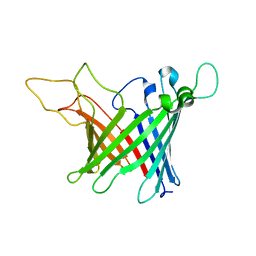 | | Solid-state NMR Structure of outer membrane protein G in lipid bilayers | | Descriptor: | Outer membrane protein G | | Authors: | Retel, J.S, Nieuwkoop, A.J, Hiller, M, Higman, V.A, Barbet-Massin, E, Stanek, J, Andreas, L.B, Franks, W.T, van Rossum, B.-J, Vinothkumar, K.R, Handel, L, de Palma, G.G, Bardiaux, B, Pintacuda, G, Emsley, L, Kuelbrandt, W, Oschkinat, H. | | Deposit date: | 2017-01-20 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure of outer membrane protein G in lipid bilayers.
Nat Commun, 8, 2017
|
|
2W80
 
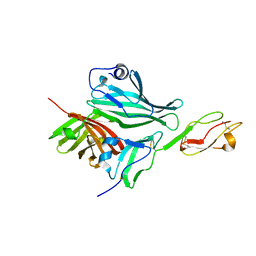 | | Structure of a complex between Neisseria meningitidis factor H binding protein and CCPs 6-7 of human complement factor H | | Descriptor: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Schneider, M.C, Prosser, B.E, Caesar, J.J.E, Kugelberg, E, Li, S, Zhang, Q, Quoraishi, S, Lovett, J.E, Deane, J.E, Sim, R.B, Roversi, P, Johnson, S, Tang, C.M, Lea, S.M. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Neisseria Meningitidis Recruits Factor H Using Protein Mimicry of Host Carbohydrates.
Nature, 458, 2009
|
|
2W81
 
 | | Structure of a complex between Neisseria meningitidis factor H binding protein and CCPs 6-7 of human complement factor H | | Descriptor: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Schneider, M.C, Prosser, B.E, Caesar, J.J.E, Kugelberg, E, Li, S, Zhang, Q, Quoraishi, S, Lovett, J.E, Deane, J.E, Sim, R.B, Roversi, P, Johnson, S, Tang, C.M, Lea, S.M. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Neisseria Meningitidis Recruits Factor H Using Protein Mimicry of Host Carbohydrates.
Nature, 458, 2009
|
|
1NLM
 
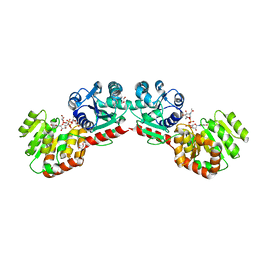 | | CRYSTAL STRUCTURE OF MURG:GLCNAC COMPLEX | | Descriptor: | GLYCEROL, UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Hu, Y, Chen, L, Ha, S, Gross, B, Falcone, B, Walker, D, Mokhtarzadeh, M, Walker, S. | | Deposit date: | 2003-01-07 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2LME
 
 | | Solid-state NMR structure of the membrane anchor domain of the trimeric autotransporter YadA | | Descriptor: | Adhesin yadA | | Authors: | Shahid, S.A, Bardiaux, B, Franks, W.T, Habeck, M, Linke, D, van Rossum, B. | | Deposit date: | 2011-11-30 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals.
Nat.Methods, 9, 2012
|
|
3LJ9
 
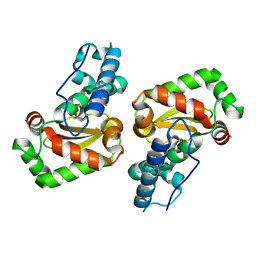 | | X-ray structure of the iron superoxide dismutase from pseudoalteromonas haloplanktis in complex with sodium azide | | Descriptor: | AZIDE ION, FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
3LIO
 
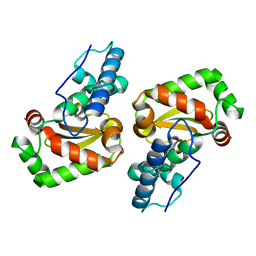 | | X-ray structure of the iron superoxide dismutase from pseudoalteromonas haloplanktis (crystal form I) | | Descriptor: | FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, iron superoxide dismutase | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-25 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
3LJF
 
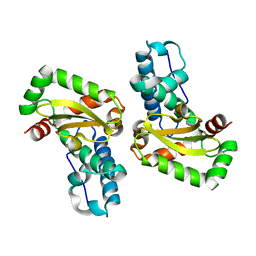 | | The X-ray structure of iron superoxide dismutase from Pseudoalteromonas haloplanktis (crystal form II) | | Descriptor: | FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, iron superoxide dismutase | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
2KLR
 
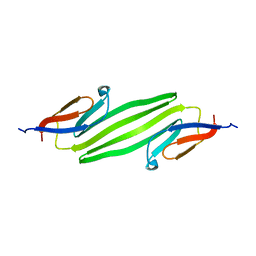 | | Solid-state NMR structure of the alpha-crystallin domain in alphaB-crystallin oligomers | | Descriptor: | Alpha-crystallin B chain | | Authors: | Jehle, S, Rajagopal, P, Markovic, S, Bardiaux, B, Kuehne, R, Higman, V.A, Klevit, R.E, van Rossum, B, Oschkinat, H. | | Deposit date: | 2009-07-08 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR and SAXS studies provide a structural basis for the activation of alphaB-crystallin oligomers.
Nat.Struct.Mol.Biol., 17, 2010
|
|
