1AIO
 
 | | CRYSTAL STRUCTURE OF A DOUBLE-STRANDED DNA CONTAINING THE MAJOR ADDUCT OF THE ANTICANCER DRUG CISPLATIN | | Descriptor: | Cisplatin, DNA (5'-D(*CP*CP*(BRU)P*CP*TP*[PT(NH3)2(GP*GP)]*TP*CP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*GP*AP*CP*CP*AP*GP*AP*GP*G)-3') | | Authors: | Takahara, P.M, Rosenzweig, A.C, Frederick, C.A, Lippard, S.J. | | Deposit date: | 1997-04-23 | | Release date: | 1997-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin.
Nature, 377, 1995
|
|
2B8E
 
 | |
2B7J
 
 | |
2ALX
 
 | | Ribonucleotide Reductase R2 from Escherichia coli in space group P6(1)22 | | Descriptor: | MANGANESE (II) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 | | Authors: | Sommerhalter, M, Saleh, L, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2005-08-08 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Escherichia coli ribonucleotide reductase R2 in space group P6122.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2B7K
 
 | |
8SR4
 
 | | particulate methane monooxygeanse treated with potassium cyanide and copper reloaded | | Descriptor: | Ammonia monooxygenase/methane monooxygenase, subunit C family protein, COPPER (II) ION, ... | | Authors: | Tucci, F.J, Jodts, R.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-05 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
8SR2
 
 | | particulate methane monooxygenase incubated with 4,4,4-trifluorobutanol | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-05 | | Release date: | 2023-11-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Product analogue binding identifies the copper active site of particulate methane monooyxgenase
Nat Catal, 2023
|
|
8SQW
 
 | | particulate methane monooxygenase crosslinked with 2,2,2-trifluoroethanol bound | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
8SR1
 
 | | particulate methane monooxygenase crosslinked with 4,4,4-trifluorobutanol bound | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, 4,4,4-trifluorobutan-1-ol, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-05 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
8SR5
 
 | |
6E1C
 
 | |
3IWX
 
 | |
3IWL
 
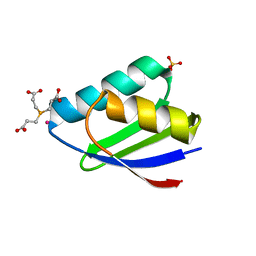 | | Crystal structure of cisplatin bound to a human copper chaperone (monomer) | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Copper transport protein ATOX1, PLATINUM (II) ION, ... | | Authors: | Boal, A.K, Rosenzweig, A.C. | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of cisplatin bound to a human copper chaperone.
J.Am.Chem.Soc., 131, 2009
|
|
2IN4
 
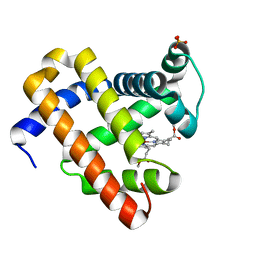 | | Crystal Structure of Myoglobin with Charge Neutralized Heme, ZnDMb-dme | | Descriptor: | Myoglobin, SULFATE ION, ZINC(II)-DEUTEROPORPHYRIN DIMETHYLESTER | | Authors: | Wheeler, K.E, Nocek, J.M, Cull, D.A, Yatsunyk, L.A, Rosenzweig, A.C, Hoffman, B.M. | | Deposit date: | 2006-10-05 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dynamic docking of cytochrome b5 with myoglobin and alpha-hemoglobin: heme-neutralization "squares" and the binding of electron-transfer-reactive configurations.
J.Am.Chem.Soc., 129, 2007
|
|
2H2M
 
 | |
2HC8
 
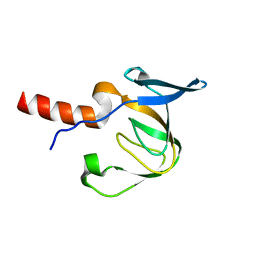 | | Structure of the A. fulgidus CopA A-domain | | Descriptor: | Cation-transporting ATPase, P-type | | Authors: | Sazinsky, M.H, Agawal, S, Arguello, J.M, Rosenzweig, A.C. | | Deposit date: | 2006-06-15 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the Actuator Domain from the Archaeoglobus fulgidus Cu(+)-ATPase(,).
Biochemistry, 45, 2006
|
|
2HU9
 
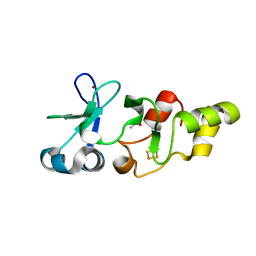 | | X-ray structure of the Archaeoglobus fulgidus CopZ N-terminal Domain | | Descriptor: | ACETIC ACID, FE2/S2 (INORGANIC) CLUSTER, Mercuric transport protein periplasmic component, ... | | Authors: | Sazinsky, M.H, LeMoine, B, Arguello, J.M, Rosenzweig, A.C. | | Deposit date: | 2006-07-26 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Characterization and structure of a Zn2+ and [2Fe-2S]-containing copper chaperone from Archaeoglobus fulgidus.
J.Biol.Chem., 282, 2007
|
|
7L6G
 
 | | MbnP from Methylosinus trichosporium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Manesis, A.C, Rosenzweig, A.C. | | Deposit date: | 2020-12-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Copper binding by a unique family of metalloproteins is dependent on kynurenine formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6P1F
 
 | | apo PmoF2 PCuAC domain | | Descriptor: | Copper chaperone PCu(A)C | | Authors: | Fisher, O.S, Sendzik, M.R, Rosenzweig, A.C. | | Deposit date: | 2019-05-19 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.654 Å) | | Cite: | PCuAC domains from methane-oxidizing bacteria use a histidine brace to bind copper.
J.Biol.Chem., 294, 2019
|
|
6P17
 
 | |
6P1E
 
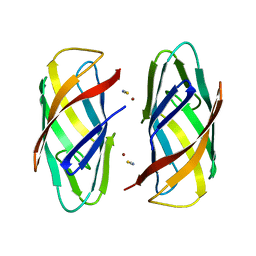 | |
6P16
 
 | |
6P1G
 
 | | Copper-bound PCuAC domain from PmoF2 | | Descriptor: | COPPER (II) ION, Copper chaperone PCu(A)C, ZINC ION | | Authors: | Fisher, O.S, Rosenzweig, A.C. | | Deposit date: | 2019-05-19 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | PCuAC domains from methane-oxidizing bacteria use a histidine brace to bind copper.
J.Biol.Chem., 294, 2019
|
|
3N3A
 
 | | Ribonucleotide Reductase Dimanganese(II)-NrdF from Escherichia coli in Complex with Reduced NrdI | | Descriptor: | FLAVIN MONONUCLEOTIDE, MANGANESE (II) ION, Protein nrdI, ... | | Authors: | Boal, A.K, Cotruvo Jr, J.A, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2010-05-19 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for activation of class Ib ribonucleotide reductase.
Science, 329, 2010
|
|
3N39
 
 | | Ribonucleotide Reductase Dimanganese(II)-NrdF from Escherichia coli in Complex with NrdI | | Descriptor: | FLAVIN MONONUCLEOTIDE, MANGANESE (II) ION, Protein nrdI, ... | | Authors: | Boal, A.K, Cotruvo Jr, J.A, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2010-05-19 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for activation of class Ib ribonucleotide reductase.
Science, 329, 2010
|
|
