1GJH
 
 | | HUMAN BCL-2, ISOFORM 2 | | Descriptor: | PROTEIN (APOPTOSIS REGULATOR BCL-2 WITH PUTATIVE FLEXIBLE LOOP REPLACED WITH A PORTION OF APOPTOSIS REGULATOR BCL-X PROTEIN) | | Authors: | Petros, A.M, Medek, A, Nettesheim, D.G, Kim, D.H, Yoon, H.S, Swift, K, Matayoshi, E.D, Oltersdorf, T, Fesik, S.W. | | Deposit date: | 2001-05-31 | | Release date: | 2001-06-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the antiapoptotic protein bcl-2.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1G5M
 
 | | HUMAN BCL-2, ISOFORM 1 | | Descriptor: | PROTEIN (APOPTOSIS REGULATOR BCL-2 WITH PUTATIVE FLEXIBLE LOOP REPLACED WITH A PORTION OF APOPTOSIS REGULATOR BCL-X PROTEIN) | | Authors: | Petros, A.M, Medek, A, Nettesheim, D.G, Kim, D.H, Yoon, H.S, Swift, K, Matayoshi, E.D, Oltersdorf, T, Fesik, S.W. | | Deposit date: | 2000-11-01 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the antiapoptotic protein bcl-2.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1GMX
 
 | | Escherichia coli GlpE sulfurtransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, THIOSULFATE SULFURTRANSFERASE GLPE | | Authors: | Spallarossa, A, Donahue, J.T, Larson, T.J, Bolognesi, M, Bordo, D. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Escherichia Coli Glpe is a Prototype Sulfurtransferase for the Single-Domain Rhodanese Homology Superfamily
Structure, 9, 2001
|
|
1GU1
 
 | | Crystal structure of type II dehydroquinase from Streptomyces coelicolor complexed with 2,3-anhydro-quinic acid | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Roszak, A.W, Robinson, D.A, Krell, T, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
1H05
 
 | |
1GTZ
 
 | | Structure of STREPTOMYCES COELICOLOR TYPE II DEHYDROQUINASE R23A MUTANT IN COMPLEX WITH DEHYDROSHIKIMATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, 3-DEHYDROSHIKIMATE | | Authors: | Roszak, A.W, Krell, T, Robinson, D.A, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
1XJL
 
 | |
5U5R
 
 | | Crystal Structure and X-ray Diffraction Data Collection of Importin-alpha from Mus musculus Complexed with a PMS2 NLS Peptide | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Importin subunit alpha-1, Mismatch repair endonuclease PMS2 | | Authors: | Barros, A.C, Takeda, A.A, Dreyer, T.R, Velazquez-Campoy, A, Kobe, B, Fontes, M.R. | | Deposit date: | 2016-12-07 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA mismatch repair proteins MLH1 and PMS2 can be imported to the nucleus by a classical nuclear import pathway.
Biochimie, 146, 2018
|
|
1GU0
 
 | | CRYSTAL STRUCTURE OF TYPE II DEHYDROQUINASE FROM STREPTOMYCES COELICOLOR | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Roszak, A.W, Krell, T, Robinson, D, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
2VG7
 
 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-iodophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
1NYT
 
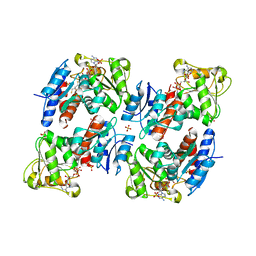 | | SHIKIMATE DEHYDROGENASE AroE COMPLEXED WITH NADP+ | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Roszak, A.W, Lapthorn, A.J. | | Deposit date: | 2003-02-13 | | Release date: | 2003-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of shikimate dehydrogenase AroE and its Paralog YdiB. A common structural framework for different activities.
J.Biol.Chem., 278, 2003
|
|
2VG5
 
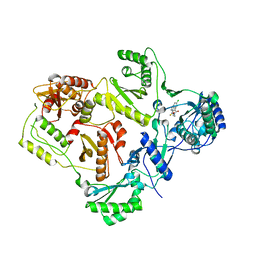 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-chlorophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
1G5J
 
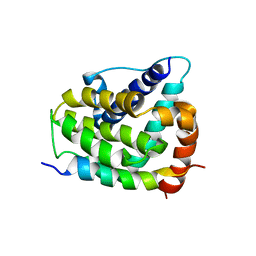 | | COMPLEX OF BCL-XL WITH PEPTIDE FROM BAD | | Descriptor: | APOPTOSIS REGULATOR BCL-X, BAD PROTEIN | | Authors: | Petros, A.M, Nettesheim, D.G, Wang, Y, Olejniczak, E.T, Meadows, R.P, Mack, J, Swift, K, Matayoshi, E.D, Zhang, H, Thompson, C.B, Fesik, S.W. | | Deposit date: | 2000-11-01 | | Release date: | 2001-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies.
Protein Sci., 9, 2000
|
|
1GN0
 
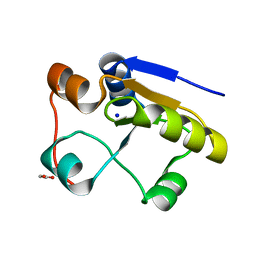 | | Escherichia coli GlpE sulfurtransferase soaked with KCN | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, THIOSULFATE SULFURTRANSFERASE GLPE | | Authors: | Spallarossa, A, Donahue, J.T, Larson, T.J, Bolognesi, M, Bordo, D. | | Deposit date: | 2001-10-01 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Escherichia Coli Glpe is a Prototype Sulfurtransferase for the Single-Domain Rhodanese Homology Superfamily
Structure, 9, 2001
|
|
2VG6
 
 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-bromophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
1MTY
 
 | | METHANE MONOOXYGENASE HYDROXYLASE FROM METHYLOCOCCUS CAPSULATUS (BATH) | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE HYDROXYLASE | | Authors: | Rosenzweig, A.C, Nordlund, P, Lippard, S.J, Frederick, C.A. | | Deposit date: | 1996-07-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): implications for substrate gating and component interactions.
Proteins, 29, 1997
|
|
1P0F
 
 | | Crystal Structure of the Binary Complex: NADP(H)-Dependent Vertebrate Alcohol Dehydrogenase (ADH8) with the cofactor NADP | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent ALCOHOL DEHYDROGENASE, ... | | Authors: | Rosell, A, Valencia, E, Pares, X, Fita, I, Farres, J, Ochoa, W.F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the vertebrate NADP(H)-dependent alcohol dehydrogenase (ADH8)
J.Mol.Biol., 330, 2003
|
|
1OOZ
 
 | | Deletion mutant of SUCCINYL-COA:3-KETOACID COA TRANSFERASE FROM PIG HEART | | Descriptor: | POTASSIUM ION, Succinyl-CoA:3-ketoacid-coenzyme A transferase | | Authors: | Coros, A.M, Swenson, L, Wolodko, W.T, Fraser, M.E. | | Deposit date: | 2003-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the CoA transferase from pig heart to 1.7 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1P0C
 
 | | Crystal Structure of the NADP(H)-Dependent Vertebrate Alcohol Dehydrogenase (ADH8) | | Descriptor: | GLYCEROL, NADP-dependent ALCOHOL DEHYDROGENASE, PHOSPHATE ION, ... | | Authors: | Rosell, A, Valencia, E, Pares, X, Fita, I, Farres, J, Ochoa, W.F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Vertebrate NADP(H)-dependent Alcohol Dehydrogenase (ADH8)
J.Mol.Biol., 330, 2003
|
|
1OPE
 
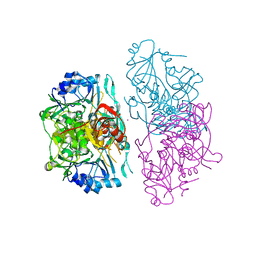 | | Deletion mutant of SUCCINYL-COA:3-KETOACID COA TRANSFERASE FROM PIG HEART | | Descriptor: | MERCURY (II) ION, POTASSIUM ION, Succinyl-CoA:3-ketoacid-coenzyme A transferase | | Authors: | Coros, A.M, Swenson, L, Wolodko, W.T, Fraser, M.E. | | Deposit date: | 2003-03-05 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the CoA transferase from pig heart to 1.7 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OOY
 
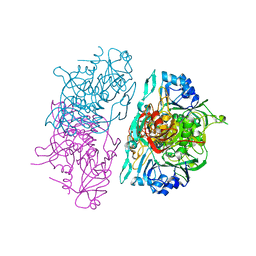 | | SUCCINYL-COA:3-KETOACID COA TRANSFERASE FROM PIG HEART | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Succinyl-CoA:3-ketoacid-coenzyme A transferase, ... | | Authors: | Coros, A.M, Swenson, L, Wolodko, W.T, Fraser, M.E. | | Deposit date: | 2003-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the CoA transferase from pig heart to 1.7 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PYH
 
 | | Crystal structure of RC-LH1 core complex from Rhodopseudomonas palustris | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Roszak, A.W, Howard, T.D, Southall, J, Gardiner, A.T, Law, C.J, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2003-07-08 | | Release date: | 2004-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Crystal structure of the RC-LH1 core complex from Rhodopseudomonas palustris.
Science, 302, 2003
|
|
3DZM
 
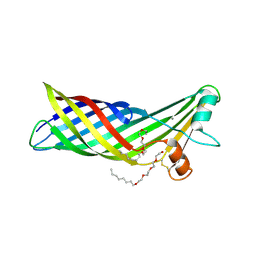 | |
4OQ6
 
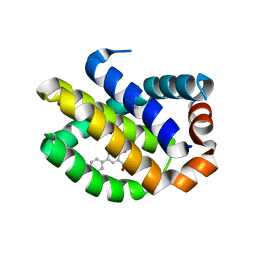 | | Crystal Structure of Human MCL-1 Bound to Inhibitor 4-hydroxy-4'-propylbiphenyl-3-carboxylic acid | | Descriptor: | 4-hydroxy-4'-propylbiphenyl-3-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Petros, A.M, Swann, S.L, Song, D, Swinger, K, Park, C, Zhang, H, Wendt, M.D, Kunzer, A.R, Souers, A.J, Sun, C. | | Deposit date: | 2014-02-07 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fragment-based discovery of potent inhibitors of the anti-apoptotic MCL-1 protein.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4OQ5
 
 | | Crystal Structure of Human MCL-1 Bound to Inhibitor 4-(4-methylnaphthalen-1-yl)-2-{[(4-phenoxyphenyl)sulfonyl]amino}benzoic acid | | Descriptor: | 4-(4-methylnaphthalen-1-yl)-2-{[(4-phenoxyphenyl)sulfonyl]amino}benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Petros, A.M, Swann, S.L, Song, D, Swinger, K, Park, C, Zhang, H, Wendt, M.D, Kunzer, A.R, Souers, A.J, Sun, C. | | Deposit date: | 2014-02-07 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Fragment-based discovery of potent inhibitors of the anti-apoptotic MCL-1 protein.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
