8DOA
 
 | |
7T2F
 
 | | Solution structure of the model HEEH mini protein homodimer HEEH_TK_rd5_0341 | | 分子名称: | HEEH mini protein HEEH_TK_rd5_0341 | | 著者 | Lemak, A, Houliston, S, Kim, T.-E, Martel, C, Rocklin, G.J, Arrowsmith, C.H. | | 登録日 | 2021-12-04 | | 公開日 | 2022-10-05 | | 最終更新日 | 2022-10-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Dissecting the stability determinants of a challenging de novo protein fold using massively parallel design and experimentation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8SHM
 
 | |
5UOI
 
 | | Solution structure of the de novo mini protein HHH_rd1_0142 | | 分子名称: | HHH_rd1_0142 | | 著者 | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | 登録日 | 2017-01-31 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
8SKX
 
 | |
8SKE
 
 | |
8SKD
 
 | |
5UP5
 
 | | Solution structure of the de novo mini protein EHEE_rd1_0284 | | 分子名称: | EHEE_rd1_0284 | | 著者 | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | 登録日 | 2017-02-01 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5UYO
 
 | | Solution NMR structure of the de novo mini protein HEEH_rd4_0097 | | 分子名称: | HEEH_rd4_0097 | | 著者 | Lemak, A, Rocklin, G.J, Houliston, S, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | 登録日 | 2017-02-24 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5UP1
 
 | | Solution structure of the de novo mini protein EEHEE_rd3_1049 | | 分子名称: | EEHEE_rd3_1049 | | 著者 | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | 登録日 | 2017-02-01 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
3HUQ
 
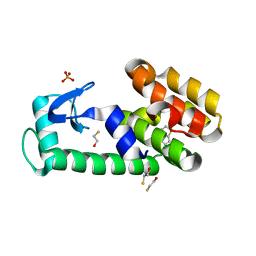 | | Thieno[3,2-b]thiophene in complex with T4 lysozyme L99A/M102Q | | 分子名称: | BETA-MERCAPTOETHANOL, Lysozyme, PHOSPHATE ION, ... | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-15 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HU8
 
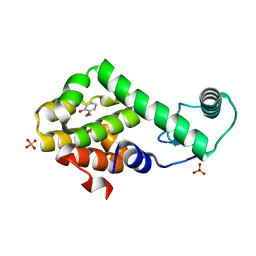 | | 2-ethoxyphenol in complex with T4 lysozyme L99A/M102Q | | 分子名称: | 2-ethoxyphenol, Lysozyme, PHOSPHATE ION | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-13 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HU9
 
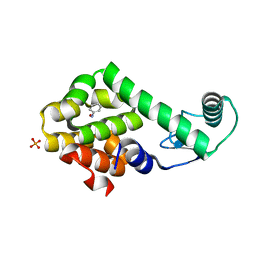 | | Nitrosobenzene in complex with T4 lysozyme L99A/M102Q | | 分子名称: | Lysozyme, NITROSOBENZENE, PHOSPHATE ION | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-13 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HT9
 
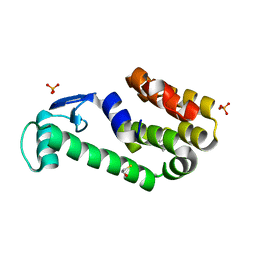 | | 2-methoxyphenol in complex with T4 lysozyme L99A/M102Q | | 分子名称: | BETA-MERCAPTOETHANOL, Guaiacol, Lysozyme, ... | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-11 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HT8
 
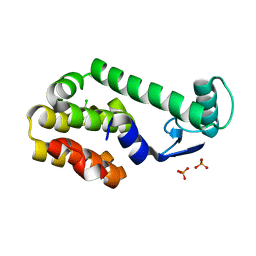 | | 5-chloro-2-methylphenol in complex with T4 lysozyme L99A/M102Q | | 分子名称: | 5-chloro-2-methylphenol, Lysozyme, PHOSPHATE ION | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-11 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTG
 
 | | 2-ethoxy-3,4-dihydro-2h-pyran in complex with T4 lysozyme L99A/M102Q | | 分子名称: | (2S)-2-ethoxy-3,4-dihydro-2H-pyran, BETA-MERCAPTOETHANOL, Lysozyme, ... | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-11 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HUA
 
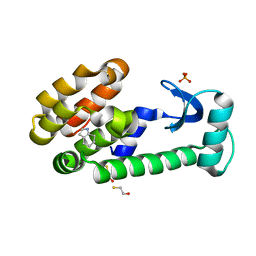 | | 4,5,6,7-tetrahydroindole in complex with T4 lysozyme L99A/M102Q | | 分子名称: | 4,5,6,7-tetrahydro-1H-indole, BETA-MERCAPTOETHANOL, Lysozyme, ... | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-13 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTB
 
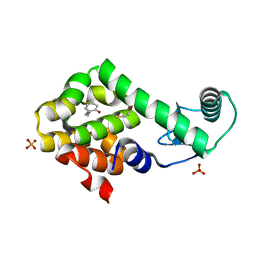 | | 2-propylphenol in complex with T4 lysozyme L99A/M102Q | | 分子名称: | 2-propylphenol, BETA-MERCAPTOETHANOL, Lysozyme, ... | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-11 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HT7
 
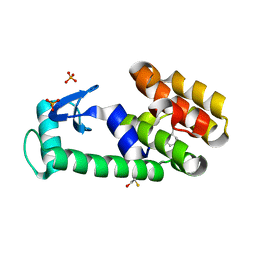 | | 2-ethylphenol in complex with T4 lysozyme L99A/M102Q | | 分子名称: | 2-ethylphenol, BETA-MERCAPTOETHANOL, Lysozyme, ... | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-11 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTF
 
 | | 4-chloro-1h-pyrazole in complex with T4 lysozyme L99A/M102Q | | 分子名称: | 4-chloro-1H-pyrazole, BETA-MERCAPTOETHANOL, Lysozyme, ... | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-11 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HT6
 
 | | 2-methylphenol in complex with T4 lysozyme L99A/M102Q | | 分子名称: | Lysozyme, PHOSPHATE ION, o-cresol | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-11 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTD
 
 | | (Z)-Thiophene-2-carboxaldoxime in complex with T4 lysozyme L99A/M102Q | | 分子名称: | (NZ)-N-(thiophen-2-ylmethylidene)hydroxylamine, BETA-MERCAPTOETHANOL, Lysozyme, ... | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-11 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HUK
 
 | | Benzylacetate in complex with T4 lysozyme L99A/M102Q | | 分子名称: | BETA-MERCAPTOETHANOL, Lysozyme, PHOSPHATE ION, ... | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-14 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
