2KB5
 
 | | Solution NMR Structure of Eosinophil Cationic Protein/RNase 3 | | Descriptor: | Eosinophil cationic protein | | Authors: | Rico, M, Bruix, M, Laurents, D.V, Santoro, J, Jimenez, M, Boix, E, Moussaoui, M, Nogues, M. | | Deposit date: | 2008-11-20 | | Release date: | 2009-06-23 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The (1)H, (13)C, (15)N resonance assignment, solution structure, and residue level stability of eosinophil cationic protein/RNase 3 determined by NMR spectroscopy
Biopolymers, 91, 2009
|
|
1TRL
 
 | | NMR SOLUTION STRUCTURE OF THE C-TERMINAL FRAGMENT 255-316 OF THERMOLYSIN: A DIMER FORMED BY SUBUNITS HAVING THE NATIVE STRUCTURE | | Descriptor: | THERMOLYSIN FRAGMENT 255 - 316 | | Authors: | Rico, M, Jimenez, M.A, Gonzalez, C, De Filippis, V, Fontana, A. | | Deposit date: | 1994-09-02 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the C-terminal fragment 255-316 of thermolysin: a dimer formed by subunits having the native structure.
Biochemistry, 33, 1994
|
|
1PNB
 
 | | STRUCTURE OF NAPIN BNIB, NMR, 10 STRUCTURES | | Descriptor: | NAPIN BNIB | | Authors: | Rico, M, Bruix, M, Gonzalez, C, Monsalve, R, Rodriguez, R. | | Deposit date: | 1996-09-17 | | Release date: | 1997-09-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | 1H NMR assignment and global fold of napin BnIb, a representative 2S albumin seed protein.
Biochemistry, 35, 1996
|
|
6ESS
 
 | | Artificial imine reductase mutant S112A-N118P-K121A-S122M | | Descriptor: | IRIDIUM ION, Streptavidin, {N-(4-{[2-(amino-kappaN)ethyl]sulfamoyl-kappaN}phenyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(chloro)[(1,2,3,4,5-eta)-1,2,3,4,5-pentamethylcyclopentadienyl]iridium(III) | | Authors: | Hestericova, M, Heinisch, T, Alonso-Cotchico, L, Marechal, J.-D, Vidossich, P, Ward, T.R. | | Deposit date: | 2017-10-24 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Directed Evolution of an Artificial Imine Reductase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6ESU
 
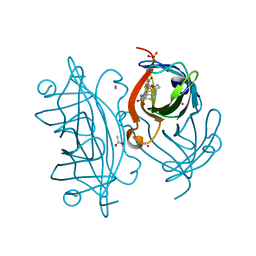 | | Artificial imine reductase mutant S112A-N118P-K121A-S122M | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[4-(2-azanylethylsulfamoyl)phenyl]pentanamide, ACETATE ION, IRIDIUM ION, ... | | Authors: | Hestericova, M, Heinisch, T, Alonso-Cotchico, L, Marechal, J.-D, Vidossich, P, Ward, T.R. | | Deposit date: | 2017-10-24 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Directed Evolution of an Artificial Imine Reductase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
2AAS
 
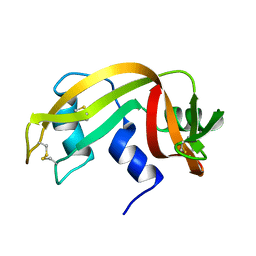 | | HIGH-RESOLUTION THREE-DIMENSIONAL STRUCTURE OF RIBONUCLEASE A IN SOLUTION BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | RIBONUCLEASE A | | Authors: | Santoro, J, Gonzalez, C, Bruix, M, Neira, J.L, Nieto, J.L, Herranz, J, Rico, M. | | Deposit date: | 1992-11-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | High-resolution three-dimensional structure of ribonuclease A in solution by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 229, 1993
|
|
1R4Y
 
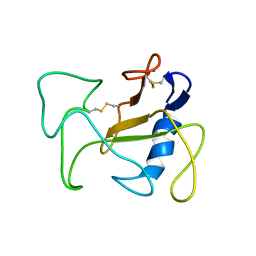 | | SOLUTION STRUCTURE OF THE DELETION MUTANT DELTA(7-22) OF THE CYTOTOXIC RIBONUCLEASE ALPHA-SARCIN | | Descriptor: | Ribonuclease alpha-sarcin | | Authors: | Garcia-Mayoral, M.F, Garcia-Ortega, L, Lillo, M.P, Santoro, J, Martinez Del Pozo, A, Gavilanes, J.G, Rico, M, Bruix, M. | | Deposit date: | 2003-10-09 | | Release date: | 2004-04-06 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the noncytotoxic {alpha}-sarcin mutant {Delta}(7-22): The importance of the native conformation of peripheral loops for activity.
Protein Sci., 13, 2004
|
|
2L3L
 
 | | The solution structure of the N-terminal domain of human Tubulin Binding Cofactor C reveals a platform for the interaction with ab-tubulin | | Descriptor: | Tubulin-specific chaperone C | | Authors: | Garcia-Mayoral, M.F, Castano, R, Lopez-Fanarraga, M.L, Zabala, J.C, Rico, M, Bruix, M. | | Deposit date: | 2010-09-14 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of human tubulin binding cofactor C reveals a platform for tubulin interaction
To be Published
|
|
1C54
 
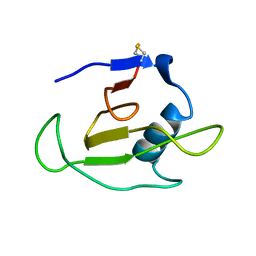 | | SOLUTION STRUCTURE OF RIBONUCLEASE SA | | Descriptor: | RIBONUCLEASE SA | | Authors: | Laurents, D.V, Canadillas-Perez, J.M, Santoro, J, Schell, D, Pace, C.N, Rico, M, Bruix, M. | | Deposit date: | 1999-10-22 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ribonuclease Sa.
Proteins, 44, 2001
|
|
1ZUG
 
 | | STRUCTURE OF PHAGE 434 CRO PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | PHAGE 434 CRO PROTEIN | | Authors: | Padmanabhan, S, Jimenez, M.A, Gonzalez, C, Sanz, J.M, Gimenez-Gallego, G, Rico, M. | | Deposit date: | 1997-03-14 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and stability of phage 434 Cro protein.
Biochemistry, 36, 1997
|
|
1RML
 
 | | NMR STUDY OF ACID FIBROBLAST GROWTH FACTOR BOUND TO 1,3,6-NAPHTHALENE TRISULPHONATE, 26 STRUCTURES | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR, NAPHTHALENE TRISULFONATE | | Authors: | Lozano, R.M, Jimenez, M.A, Santoro, J, Rico, M, Gimenez-Gallego, G. | | Deposit date: | 1998-05-21 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of acidic fibroblast growth factor bound to 1,3, 6-naphthalenetrisulfonate: a minimal model for the anti-tumoral action of suramins and suradistas.
J.Mol.Biol., 281, 1998
|
|
1N96
 
 | | DIMERIC SOLUTION STRUCTURE OF THE CYCLIC OCTAMER CD(CGCTCATT) | | Descriptor: | CYCLIC OLIGONUCLEOTIDE D(CGCTCATT) | | Authors: | Escaja, N, Gelpi, J.L, Orozco, M, Rico, M, Pedroso, E, Gonzalez, C. | | Deposit date: | 2002-11-22 | | Release date: | 2003-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Four-stranded DNA structure stabilized by a novel G:C:A:T tetrad.
J.Am.Chem.Soc., 125, 2003
|
|
1AFP
 
 | | SOLUTION STRUCTURE OF THE ANTIFUNGAL PROTEIN FROM ASPERGILLUS GIGANTEUS. EVIDENCE FOR DISULPHIDE CONFIGURATIONAL ISOMERISM | | Descriptor: | ANTIFUNGAL PROTEIN FROM ASPERGILLUS GIGANTEUS | | Authors: | Campos-Olivas, R, Bruix, M, Santoro, J, Lacadena, J, Del Pozo, A.M, Gavilanes, J.G, Rico, M. | | Deposit date: | 1994-11-11 | | Release date: | 1995-02-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the antifungal protein from Aspergillus giganteus: evidence for cysteine pairing isomerism.
Biochemistry, 34, 1995
|
|
1DE3
 
 | | SOLUTION STRUCTURE OF THE CYTOTOXIC RIBONUCLEASE ALPHA-SARCIN | | Descriptor: | RIBONUCLEASE ALPHA-SARCIN | | Authors: | Perez-Canadillas, J.M, Campos-Olivas, R, Santoro, J, Lacadena, J, Martinez del Pozo, A, Gavilanes, J.G, Rico, M, Bruix, M. | | Deposit date: | 1999-11-12 | | Release date: | 2000-06-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The highly refined solution structure of the cytotoxic ribonuclease alpha-sarcin reveals the structural requirements for substrate recognition and ribonucleolytic activity.
J.Mol.Biol., 299, 2000
|
|
1E68
 
 | | Solution structure of bacteriocin AS-48 | | Descriptor: | AS-48 PROTEIN | | Authors: | Gonzalez, C, Langdon, G, Bruix, M, Galvez, A, Valdivia, E, Maqueda, M, Rico, M. | | Deposit date: | 2000-08-09 | | Release date: | 2000-10-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Bacteriocin as-48, a Microbial Cyclic Polypeptide Structurally and Functionally Related to Mammalian Nk-Lysin
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1CYE
 
 | | THREE DIMENSIONAL STRUCTURE OF CHEMOTACTIC CHE Y PROTEIN IN AQUEOUS SOLUTION BY NUCLEAR MAGNETIC RESONANCE METHODS | | Descriptor: | CHEY | | Authors: | Santoro, J, Bruix, M, Pascual, J, Lopez, E, Serrano, L, Rico, M. | | Deposit date: | 1994-10-21 | | Release date: | 1995-02-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of chemotactic Che Y protein in aqueous solution by nuclear magnetic resonance methods.
J.Mol.Biol., 247, 1995
|
|
1SS3
 
 | | Solution structure of Ole e 6, an allergen from olive tree pollen | | Descriptor: | Pollen allergen Ole e 6 | | Authors: | Trevino, M.A, Garcia-Mayoral, M.F, Barral, P, Villalba, M, Santoro, J, Rico, M, Rodriguez, R, Bruix, M. | | Deposit date: | 2004-03-23 | | Release date: | 2004-08-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Ole e 6, a Major Allergen from Olive Tree Pollen.
J.Biol.Chem., 279, 2004
|
|
1SM7
 
 | | Solution structure of the recombinant pronapin precursor, BnIb. | | Descriptor: | recombinant Ib pronapin | | Authors: | Pantoja-Uceda, D, Palomares, O, Bruix, M, Villalba, M, Rodriguez, R, Rico, M, Santoro, J. | | Deposit date: | 2004-03-08 | | Release date: | 2005-02-01 | | Last modified: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability against digestion of rproBnIb, a recombinant 2S albumin from rapeseed: relationship to its allergenic properties.
Biochemistry, 43, 2004
|
|
1EU2
 
 | |
1EU6
 
 | |
1S6D
 
 | | Structure in solution of a methionine-rich 2S Albumin protein from Sunflower Seed | | Descriptor: | Albumin 8 | | Authors: | Pantoja-Uceda, D, Bruix, M, Shewry, P.R, Santoro, J, Rico, M. | | Deposit date: | 2004-01-23 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Methionine-Rich 2S Albumin from Sunflower Seeds: Relationship to Its Allergenic and Emulsifying Properties.
Biochemistry, 43, 2004
|
|
1PSY
 
 | | STRUCTURE OF RicC3, NMR, 20 STRUCTURES | | Descriptor: | 2S albumin | | Authors: | Pantoja-Uceda, D, Bruix, M, Gimenez-Gallego, G, Rico, M, Santoro, J. | | Deposit date: | 2003-06-22 | | Release date: | 2004-01-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RicC3, a 2S albumin storage protein from Ricinus communis.
Biochemistry, 42, 2003
|
|
1DZC
 
 | | High resolution structure of acidic fibroblast growth factor. Mutant FGF-4-ALA-(24-154), 24 NMR structures | | Descriptor: | FIBROBLAST GROWTH FACTOR 1 | | Authors: | Lozano, R.M, Pineda-Lucena, A, Gonzalez, C, Jimenez, M.A, Cuevas, P, Redondo-Horcajo, M, Sanz, J.M, Rico, M, Gimenez-Gallego, G. | | Deposit date: | 2000-02-24 | | Release date: | 2000-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H-NMR Structural Characterization of a Non Mitogenic, Vasodilatory, Ischemia-Protector and Neuromodulatory Acidic Fibroblast Growth Factor
Biochemistry, 39, 2000
|
|
1DZD
 
 | | High resolution structure of acidic fibroblast growth factor (27-154), 24 NMR structures | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Lozano, R.M, Pineda-Lucena, A, Gonzalez, C, Jimenez, M.A, Cuevas, P, Redondo-Horcajo, M, Sanz, J.M, Rico, M, Gimenez-Gallego, G. | | Deposit date: | 2000-02-24 | | Release date: | 2000-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H-NMR Structural Characterization of a Non Mitogenic, Vasodilatory, Ischemia-Protector and Neuromodulatory Acidic Fibroblast Growth Factor
Biochemistry, 39, 2000
|
|
1GPS
 
 | | SOLUTION STRUCTURE OF GAMMA 1-H AND GAMMA 1-P THIONINS FROM BARLEY AND WHEAT ENDOSPERM DETERMINED BY 1H-NMR: A STRUCTURAL MOTIF COMMON TO TOXIC ARTHROPOD PROTEINS | | Descriptor: | GAMMA-1-P THIONIN | | Authors: | Bruix, M, Jimenez, M.A, Santoro, J, Gonzalez, C, Colilla, F.J, Mendez, E, Rico, M. | | Deposit date: | 1992-07-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of gamma 1-H and gamma 1-P thionins from barley and wheat endosperm determined by 1H-NMR: a structural motif common to toxic arthropod proteins.
Biochemistry, 32, 1993
|
|
