6UYT
 
 | | Crystal structure of K39-acetylated SUMO1 in complex with phosphorylated PML-SIM | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6UYZ
 
 | | Crystal structure of K46-acetylated SUMO1 in complex with phosphorylated DAXX | | Descriptor: | Small ubiquitin-related modifier 1, phosphorylated DAXX | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6UYV
 
 | | Crystal structure of K46-acetylated SUMO1 in complex with phosphorylated PML-SIM | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6V7R
 
 | | Crystal structure of K37-acetylated SUMO1 in complex with PIAS-SIM2 | | Descriptor: | Protein PIAS, Small ubiquitin-related modifier 1 | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-12-09 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Characterization of a C-Terminal SUMO-Interacting Motif Present in Select PIAS-Family Proteins.
Structure, 28, 2020
|
|
6UYS
 
 | | Crystal structure of K37-acetylated SUMO1 in complex with phosphorylated PML-SIM | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
6V7P
 
 | | Crystal structure of SUMO1 in complex with PIAS-SIM2 | | Descriptor: | Protein PIAS, Small ubiquitin-related modifier 1 | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-12-09 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Characterization of a C-Terminal SUMO-Interacting Motif Present in Select PIAS-Family Proteins.
Structure, 28, 2020
|
|
4HNC
 
 | |
4M6U
 
 | |
2NT8
 
 | | ATP bound at the active site of a PduO type ATP:co(I)rrinoid adenosyltransferase from Lactobacillus reuteri | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cobalamin adenosyltransferase, GLYCEROL, ... | | Authors: | St-Maurice, M, Mera, P.E, Taranto, M.P, Sesma, F, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2006-11-07 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural characterization of the active site of the PduO-type ATP:Co(I)rrinoid adenosyltransferase from Lactobacillus reuteri.
J.Biol.Chem., 282, 2007
|
|
5SXP
 
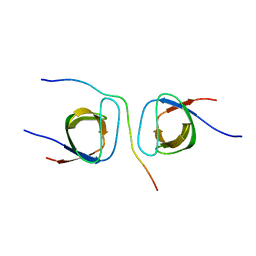 | | STRUCTURAL BASIS FOR THE INTERACTION BETWEEN ITCH PRR AND BETA-PIX | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Rho guanine nucleotide exchange factor 7 | | Authors: | Cappadocia, L, Desrochers, G, Lussier-Price, M, Angers, A, Omichinski, J.G. | | Deposit date: | 2016-08-09 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis of interactions between SH3 domain-containing proteins and the proline-rich region of the ubiquitin ligase Itch.
J. Biol. Chem., 292, 2017
|
|
4M6V
 
 | |
4MFD
 
 | |
4LOC
 
 | |
4MFE
 
 | |
4MIM
 
 | |
4FP1
 
 | | P. putida mandelate racemase co-crystallized with 3,3,3-trifluoro-2-hydroxy-2-(trifluoromethyl) propionic acid | | Descriptor: | 3,3,3-trifluoro-2-hydroxy-2-(trifluoromethyl)propanoic acid, MAGNESIUM ION, Mandelate racemase | | Authors: | Lietzan, A.D, St.Maurice, M. | | Deposit date: | 2012-06-21 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Potent inhibition of mandelate racemase by a fluorinated substrate-product analogue with a novel binding mode.
Biochemistry, 53, 2014
|
|
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | Descriptor: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | Deposit date: | 2017-03-23 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
1U9Q
 
 | | Crystal structure of cruzain bound to an alpha-ketoester | | Descriptor: | [1-(1-METHYL-4,5-DIOXO-PENT-2-ENYLCARBAMOYL)-2-PHENYL-ETHYL]-CARBAMIC ACID BENZYL ESTER, cruzipain | | Authors: | Lange, M, Weston, S.G, Cheng, H, Culliane, M, Fiorey, M.M, Grisostomi, C, Hardy, L.W, Hartstough, D.S, Pallai, P.V, Tilton, R.F, Baldino, C.M, Brinen, L.S, Engel, J.C, Choe, Y, Price, M.S, Craik, C.S. | | Deposit date: | 2004-08-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of alpha-keto-based inhibitors of cruzain, a cysteine protease implicated in Chagas disease
Bioorg.Med.Chem., 13, 2005
|
|
2MKR
 
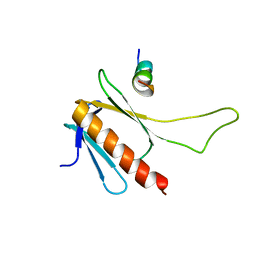 | | Structural Characterization of a Complex Between the Acidic Transactivation Domain of EBNA2 and the Tfb1/p62 subunit of TFIIH. | | Descriptor: | Epstein-Barr nuclear antigen 2, RNA polymerase II transcription factor B subunit 1 | | Authors: | Chabot, P.R, Raiola, L, Lussier-Price, M, Morse, T, Arseneault, G, Archambault, J, Omichinski, J. | | Deposit date: | 2014-02-12 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of a Complex between the Acidic Transactivation Domain of EBNA2 and the Tfb1/p62 Subunit of TFIIH.
Plos Pathog., 10, 2014
|
|
5L7R
 
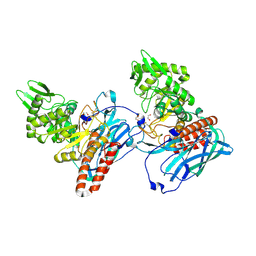 | | Crystal structure of BvGH123 | | Descriptor: | 1,2-ETHANEDIOL, glycoside hydrolase | | Authors: | Roth, C, Petricevic, M, John, A, Goddard-Borger, E.D, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-22 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and mechanistic insights into a Bacteroides vulgatus retaining N-acetyl-beta-galactosaminidase that uses neighbouring group participation.
Chem. Commun. (Camb.), 52, 2016
|
|
6VIM
 
 | | P. putida mandelate racemase co-crystallized with phenylboronic acid | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Mandelate racemase, ... | | Authors: | Grandinetti, L, Sharma, A.N, Bearne, S.L, St Maurice, M. | | Deposit date: | 2020-01-13 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent Inhibition of Mandelate Racemase by Boronic Acids: Boron as a Mimic of a Carbon Acid Center.
Biochemistry, 59, 2020
|
|
5JTS
 
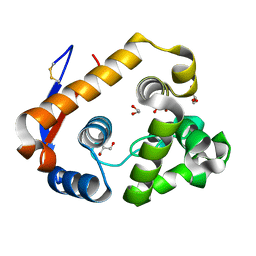 | | Structure of a beta-1,4-mannanase, SsGH134. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2016-05-09 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
5JUG
 
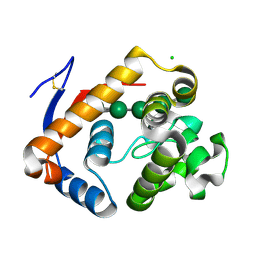 | | Structure of an inactive (E45Q) variant of a beta-1,4-mannanase, SsGH134, in complex with Man5 | | Descriptor: | CHLORIDE ION, GLYCEROL, alpha-D-mannopyranose, ... | | Authors: | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2016-05-10 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
5JU9
 
 | | Structure of a beta-1,4-mannanase, SsGH134, in complex with Man3. | | Descriptor: | CHLORIDE ION, beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2016-05-10 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
6FAR
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-mannoimidazole | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, Glycosyl hydrolase family 71, alpha-D-mannopyranose | | Authors: | Fernandes, P.Z, Petricevic, M, Sobala, L.F, Davies, G.J, Williams, S.J. | | Deposit date: | 2017-12-16 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Exploration of Strategies for Mechanism-Based Inhibitor Design for Family GH99 endo-alpha-1,2-Mannanases.
Chemistry, 24, 2018
|
|
