6OMB
 
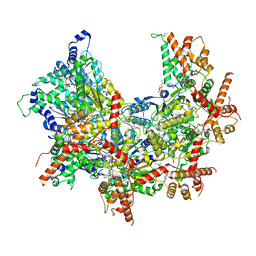 | | Cdc48 Hexamer (Subunits A to E) with substrate bound to the central pore | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Cooney, I, Han, H, Stewart, M, Carson, R.H, Hansen, D, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2019-04-18 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Cdc48 segregase in the act of unfolding an authentic substrate.
Science, 365, 2019
|
|
6OVU
 
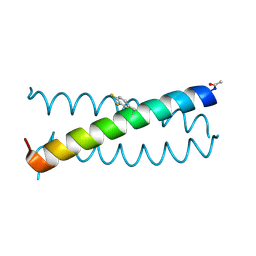 | | Coiled-coil Trimer with Glu:3,4-difluorophenylalanine:Lys Triad | | Descriptor: | Coiled-coil Trimer with Glu:3,4-difluorophenylalanine:Lys Triad | | Authors: | Smith, M.S, Stern, K.L, Billings, W.M, Price, J.L. | | Deposit date: | 2019-05-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Context-Dependent Stabilizing Interactions among Solvent-Exposed Residues along the Surface of a Trimeric Helix Bundle.
Biochemistry, 59, 2020
|
|
6O2E
 
 | |
6O2F
 
 | |
6OS8
 
 | |
6V5J
 
 | |
6V57
 
 | |
6V5I
 
 | | Coiled-coil Trimer with Ala:Leu:Ala Triad | | Descriptor: | Coiled-coil Trimer with Ala:Leu:Ala Triad, PENTAETHYLENE GLYCOL | | Authors: | Smith, M.S, Stern, K.L, Billings, W.M, Price, J.L. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Context-Dependent Stabilizing Interactions among Solvent-Exposed Residues along the Surface of a Trimeric Helix Bundle.
Biochemistry, 59, 2020
|
|
5UXT
 
 | | Coiled-coil Trimer with Glu:Trp:Lys Triad | | Descriptor: | GLYCEROL, coiled-coil trimer with Glu:Trp:Lys triad | | Authors: | Smith, M.S, Billings, W.M, Whitby, F.G, Miller, M.B, Price, J.L. | | Deposit date: | 2017-02-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Enhancing a long-range salt bridge with intermediate aromatic and nonpolar amino acids.
Org. Biomol. Chem., 15, 2017
|
|
8UAA
 
 | | Cdc48-Shp1 unfolding native substrate, Class 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-20 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8U7T
 
 | | Substrate-bound Cdc48, Class 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Lin, H.J.L, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-15 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8U9Z
 
 | | Cdc48-Shp1 unfolding native substrate, Class 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-20 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8U8I
 
 | | Cdc48-Shp1 unfolding native substrate, Class 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-18 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8U9C
 
 | | Cdc48-Shp1 unfolding native substrate, Class 5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-19 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8UA1
 
 | | Cdc48-Shp1 unfolding native substrate, Class 9 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-20 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8UB4
 
 | | Cdc48-Shp1 unfolding native substrate, consensus structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-22 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8UA0
 
 | | Cdc48-Shp1 unfolding native substrate, Class 8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-20 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8U9P
 
 | | Cdc48-Shp1 unfolding native substrate, Class 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-19 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8U9Q
 
 | | Cdc48-Shp1 unfolding native substrate, Class 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-19 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
3C3H
 
 | | alpha/beta-Peptide helix bundles: A GCN4-pLI analogue with an (alpha-alpha-beta) backbone and cyclic beta residues | | Descriptor: | alpha/beta-peptide based on the GCN4-pLI side chain sequence, with an (alpha-alpha-beta) backbone and cyclic beta-residues at positions 1, 4, ... | | Authors: | Horne, W.S, Price, J.L, Gellman, S.H. | | Deposit date: | 2008-01-28 | | Release date: | 2008-06-17 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interplay among side chain sequence, backbone composition, and residue rigidification in polypeptide folding and assembly.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C3G
 
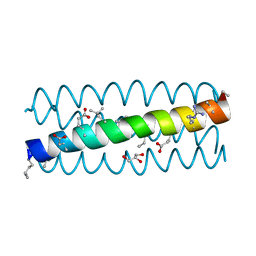 | | alpha/beta-Peptide helix bundles: The GCN4-pLI side chain sequence on an (alpha-alpha-beta) backbone | | Descriptor: | GLYCEROL, alpha/beta peptide with the GCN4-pLI side chain sequence on an (alpha-alpha-beta) backbone | | Authors: | Horne, W.S, Price, J.L, Gellman, S.H. | | Deposit date: | 2008-01-28 | | Release date: | 2008-06-17 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interplay among side chain sequence, backbone composition, and residue rigidification in polypeptide folding and assembly.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C3F
 
 | |
2M9J
 
 | | NMR solution structure of Pin1 WW domain mutant 6-1g | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-10 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M9F
 
 | | NMR solution structure of Pin1 WW domain mutant 5-1g | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-07 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M9E
 
 | | NMR solution structure of Pin1 WW domain mutant 5-1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-07 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
