6B6X
 
 | | Crystal structure of Desulfovibrio vulgaris carbon monoxide dehydrogenase, dithionite-reduced (protein batch 2), canonical C-cluster | | Descriptor: | CHLORIDE ION, Carbon monoxide dehydrogenase, FE(4)-NI(1)-S(4) CLUSTER, ... | | Authors: | Wittenborn, E.C, Drennan, C.L. | | Deposit date: | 2017-10-03 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Redox-dependent rearrangements of the NiFeS cluster of carbon monoxide dehydrogenase.
Elife, 7, 2018
|
|
3I04
 
 | | Cyanide-bound structure of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase from Moorella thermoacetica, cyanide-bound C-cluster | | Descriptor: | ACETATE ION, COPPER (I) ION, CYANIDE ION, ... | | Authors: | Kung, Y, Doukov, T.I, Drennan, C.L. | | Deposit date: | 2009-06-24 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic snapshots of cyanide- and water-bound C-clusters from bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase.
Biochemistry, 48, 2009
|
|
3EPT
 
 | |
6V59
 
 | |
6B9T
 
 | |
6B9S
 
 | |
6VUE
 
 | | wild-type choline TMA lyase in complex with 1-methyl-1,2,3,6-tetrahydropyridin-3-ol | | Descriptor: | (3S)-1-methyl-1,2,3,6-tetrahydropyridin-3-ol, Choline trimethylamine-lyase, SODIUM ION | | Authors: | Ortega, M.A, Drennan, C.L. | | Deposit date: | 2020-02-15 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of a Cyclic Choline Analog That Inhibits Anaerobic Choline Metabolism by Human Gut Bacteria.
Acs Med.Chem.Lett., 11, 2020
|
|
8STA
 
 | |
6CIQ
 
 | |
6CIO
 
 | | Pyruvate:ferredoxin oxidoreductase from Moorella thermoacetica with lactyl-TPP bound | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Chen, P.Y.-T, Drennan, C.L. | | Deposit date: | 2018-02-24 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Binding site for coenzyme A revealed in the structure of pyruvate:ferredoxin oxidoreductase fromMoorella thermoacetica.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CIP
 
 | |
6CIN
 
 | |
4TVW
 
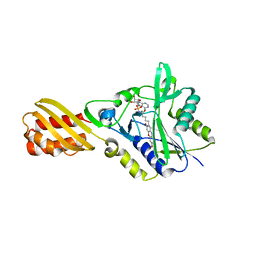 | | Resorufin ligase with bound resorufin-AMP analog | | Descriptor: | 5'-O-{[5-(7-hydroxy-3-oxo-3H-phenoxazin-2-yl)pentanoyl]sulfamoyl}adenosine, Lipoate-protein ligase A | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2014-06-28 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.505 Å) | | Cite: | Computational design of a red fluorophore ligase for site-specific protein labeling in living cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3I01
 
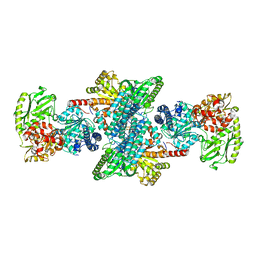 | | Native structure of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase from Moorella thermoacetica, water-bound C-cluster. | | Descriptor: | ACETATE ION, COPPER (I) ION, Carbon monoxide dehydrogenase/acetyl-CoA synthase subunit alpha, ... | | Authors: | Kung, Y, Doukov, T.I, Drennan, C.L. | | Deposit date: | 2009-06-24 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic snapshots of cyanide- and water-bound C-clusters from bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase.
Biochemistry, 48, 2009
|
|
1ELY
 
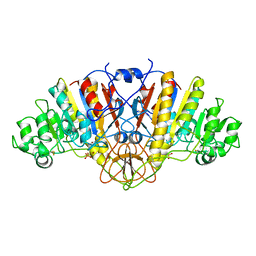 | | E. COLI ALKALINE PHOSPHATASE MUTANT (S102C) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Stec, B, Hehir, M, Brennan, C, Nolte, M, Kantrowitz, E.R. | | Deposit date: | 1998-02-10 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Kinetic and X-ray structural studies of three mutant E. coli alkaline phosphatases: insights into the catalytic mechanism without the nucleophile Ser102.
J.Mol.Biol., 277, 1998
|
|
1ELX
 
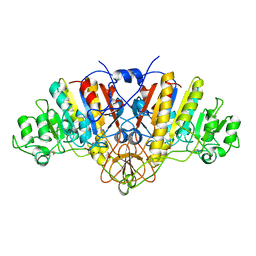 | | E. COLI ALKALINE PHOSPHATASE MUTANT (S102A) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Stec, B, Hehir, M, Brennan, C, Nolte, M, Kantrowitz, E.R. | | Deposit date: | 1998-02-10 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Kinetic and X-ray structural studies of three mutant E. coli alkaline phosphatases: insights into the catalytic mechanism without the nucleophile Ser102.
J.Mol.Biol., 277, 1998
|
|
6VXC
 
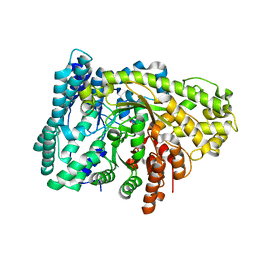 | |
6W4X
 
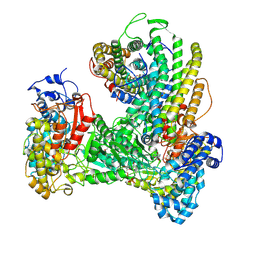 | | Holocomplex of E. coli class Ia ribonucleotide reductase with GDP and TTP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MU-OXO-DIIRON, ... | | Authors: | Kang, G, Taguchi, A, Stubbe, J, Drennan, C. | | Deposit date: | 2020-03-11 | | Release date: | 2020-04-08 | | Last modified: | 2020-05-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a trapped radical transfer pathway within a ribonucleotide reductase holocomplex.
Science, 368, 2020
|
|
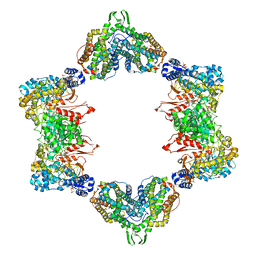
4ERM
 
 | |
3GJA
 
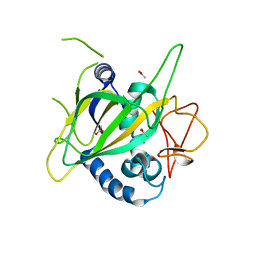 | | CytC3 | | Descriptor: | ACETATE ION, CytC3 | | Authors: | Wong, C, Drennan, C.L. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of an open active site conformation of nonheme iron halogenase CytC3
J.Am.Chem.Soc., 131, 2009
|
|
2HZV
 
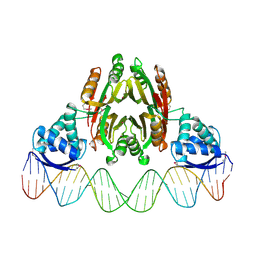 | | NikR-operator DNA complex | | Descriptor: | 5'-D(*AP*GP*TP*AP*TP*GP*AP*CP*GP*AP*AP*TP*AP*CP*TP*TP*AP*AP*AP*AP*TP*CP*GP*TP*CP*AP*TP*AP*CP*T)-3', 5'-D(*AP*GP*TP*AP*TP*GP*AP*CP*GP*AP*TP*TP*TP*TP*AP*AP*GP*TP*AP*TP*TP*CP*GP*TP*CP*AP*TP*AP*CP*T)-3', NICKEL (II) ION, ... | | Authors: | Schreiter, E.R, Drennan, C.L. | | Deposit date: | 2006-08-09 | | Release date: | 2006-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | NikR-operator complex structure and the mechanism of repressor activation by metal ions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HZA
 
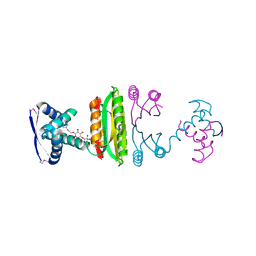 | | Nickel-bound full-length Escherichia coli NikR | | Descriptor: | 3-CYCLOHEXYLPROPYL 4-O-ALPHA-D-GLUCOPYRANOSYL-BETA-D-GLUCOPYRANOSIDE, NICKEL (II) ION, Nickel-responsive regulator | | Authors: | Schreiter, E.R, Drennan, C.L. | | Deposit date: | 2006-08-08 | | Release date: | 2006-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NikR-operator complex structure and the mechanism of repressor activation by metal ions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
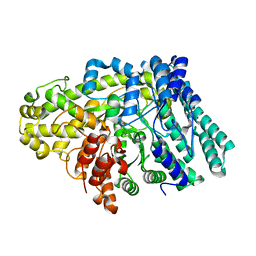
6VXE
 
 | |
4TVY
 
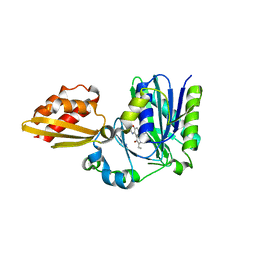 | | Apo resorufin ligase | | Descriptor: | 5-(3,7-dihydroxy-10H-phenoxazin-2-yl)pentanamide, Lipoate-protein ligase A | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2014-06-28 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Computational design of a red fluorophore ligase for site-specific protein labeling in living cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6XN6
 
 | |
