5HK3
 
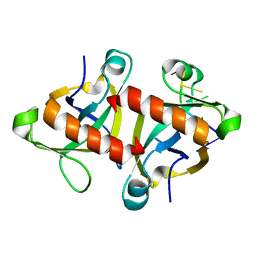 | |
6FZ5
 
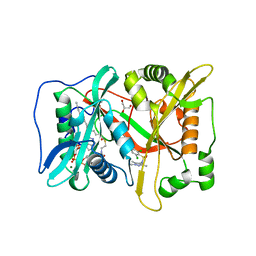 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA and inhibitor bound | | Descriptor: | 4-[3-[(8~{a}~{R})-3,4,6,7,8,8~{a}-hexahydro-1~{H}-pyrrolo[1,2-a]pyrazin-2-yl]propyl]-2,6-bis(chloranyl)-~{N}-methyl-~{N}-(1,3,5-trimethylpyrazol-4-yl)benzenesulfonamide, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Kersten, F.C, Brenk, R. | | Deposit date: | 2018-03-14 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | How To Design Selective Ligands for Highly Conserved Binding Sites: A Case Study UsingN-Myristoyltransferases as a Model System.
J.Med.Chem., 63, 2020
|
|
8QAA
 
 | |
8QAE
 
 | | X-ray crystal structure of a de novo designed single-chain antiparallel 6-helix alpha-helical barrel, sc-apCC-6-SLLA | | Descriptor: | 2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-(2-hydroxyethyloxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethanol, PENTAETHYLENE GLYCOL, sc-apCC-6-SLLA | | Authors: | Albanese, K.I, Petrenas, R, Woolfson, D.N. | | Deposit date: | 2023-08-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rationally seeded computational protein design of ɑ-helical barrels.
Nat.Chem.Biol., 20, 2024
|
|
4ZDD
 
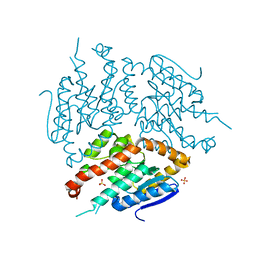 | | Structure of yeast D3,D2-enoyl-CoA isomerase bound to sulphate ion | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, SULFATE ION | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of yeast peroxisomal Delta (3), Delta (2)-enoyl-CoA isomerase complexed with acyl-CoA substrate analogues: the importance of hydrogen-bond networks for the reactivity of the catalytic base and the oxyanion hole.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ZDB
 
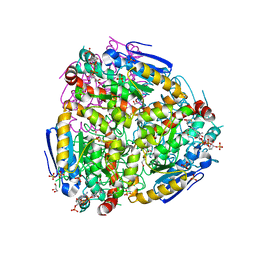 | | Yeast enoyl-CoA isomerase (ScECI2) complexed with acetoacetyl-CoA | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, ACETOACETYL-COENZYME A, GLYCEROL, ... | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of yeast peroxisomal Delta (3), Delta (2)-enoyl-CoA isomerase complexed with acyl-CoA substrate analogues: the importance of hydrogen-bond networks for the reactivity of the catalytic base and the oxyanion hole.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4DDC
 
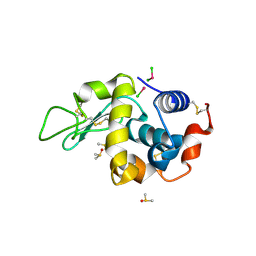 | | EVAL processed HEWL, cisplatin DMSO NAG silicone oil | | Descriptor: | Cisplatin, DIMETHYL SULFOXIDE, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6GNV
 
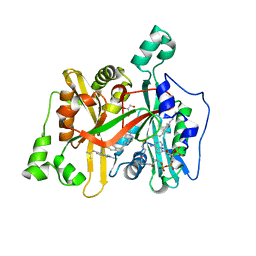 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and a isopropyl methyl indole aryl sulphonamide ligand | | Descriptor: | GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, ... | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
4DD7
 
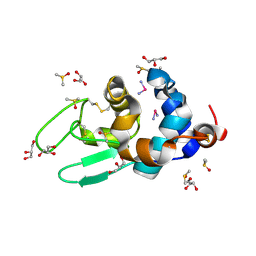 | | EVAL processed HEWL, carboplatin DMSO glycerol | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Lysozyme C, ... | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4ZDF
 
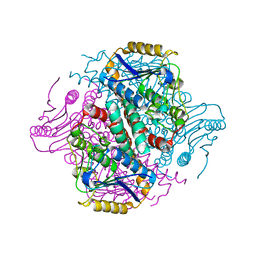 | | Crystal structure of yeast enoyl-CoA isomerase helix-10 deletion (ScECI2-H10) mutant | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, GLYCEROL | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structures of yeast peroxisomal Delta (3), Delta (2)-enoyl-CoA isomerase complexed with acyl-CoA substrate analogues: the importance of hydrogen-bond networks for the reactivity of the catalytic base and the oxyanion hole.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6GNH
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and an Azepanyl Phenyl Benzylsulphonamide Ligand | | Descriptor: | Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, methyl 4-(azepan-1-yl)-3-[(4-methoxyphenyl)sulfonylamino]benzoate | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
6GNS
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and an Azepanyl Phenyl Benzylsulphonamide Ligand | | Descriptor: | Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, methyl 4-(azepan-1-yl)-3-[[4-[4-(1-methylpiperidin-4-yl)butyl]phenyl]sulfonylamino]benzoate | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
5AG4
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A THIAZOLIDINONE LIGAND | | Descriptor: | (2S)-3-(3-chlorophenyl)-2-(pyridin-2-yl)-1,3-thiazolidin-4-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
4DD2
 
 | | EVAL processed HEWL, carboplatin aqueous glycerol | | Descriptor: | GLYCEROL, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5AGE
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A BENZOMORPHOLINONE LIGAND | | Descriptor: | 4-[(5-methyl-1,2-oxazol-3-yl)methyl]-7-[4-(1-methylpiperidin-4-yl)butyl]-2H-1,4-benzoxazin-3(4H)-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
6GNU
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and a Quionlinyl aryl sulphonamide ligand | | Descriptor: | 4-[4-(1-methylpiperidin-4-yl)butyl]-~{N}-[6-(2-methylpropyl)quinolin-5-yl]benzenesulfonamide, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
5AB6
 
 | |
4ZDE
 
 | | Crystal structure of yeast D3,D2-enoyl-CoA isomerase F268A mutant | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, GLYCEROL, SULFATE ION | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of yeast peroxisomal Delta (3), Delta (2)-enoyl-CoA isomerase complexed with acyl-CoA substrate analogues: the importance of hydrogen-bond networks for the reactivity of the catalytic base and the oxyanion hole.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4DDA
 
 | | EVAL processed HEWL, NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DD9
 
 | | EVAL processed HEWL, carboplatin DMSO paratone | | Descriptor: | DIMETHYL SULFOXIDE, Lysozyme C, carboplatin | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5AB7
 
 | |
4DD3
 
 | | EVAL processed HEWL, carboplatin aqueous paratone | | Descriptor: | 2-methylprop-1-ene, CHLORIDE ION, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6GNT
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and a Quionlinyl aryl sulphonamide ligand | | Descriptor: | 4-[4-(1-methylpiperidin-4-yl)butyl]-~{N}-(6-pyrrolidin-1-ylquinolin-5-yl)benzenesulfonamide, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, ... | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
4DDB
 
 | | EVAL processed HEWL, cisplatin DMSO paratone pH 6.5 | | Descriptor: | Cisplatin, DIMETHYL SULFOXIDE, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4ZRC
 
 | | Crystal structure of MSM-13, a putative T1-like thiolase from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase | | Authors: | Janardan, N, Harijan, R.K, Keima, T.R, Wierenga, R, Murthy, M.R.N. | | Deposit date: | 2015-05-12 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
