7O4T
 
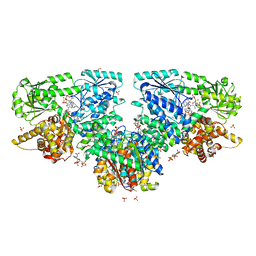 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme with Coenzyme A bound at the hydratase, thiolase active sites and possible additional binding site (CoA(ECH/HAD)) | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, 3-hydroxyacyl-CoA dehydrogenase, COENZYME A, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-04-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O4R
 
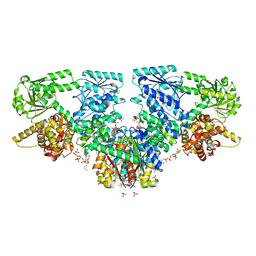 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme with Coenzyme A bound at the thiolase active sites and additional binding site (CoA(HAD/KAT)) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, COENZYME A, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-04-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O4S
 
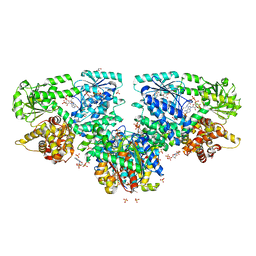 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme with Coenzyme A bound at the hydratase, thiolase active sites and additional binding site (CoA(ECH2)) | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, 3-hydroxyacyl-CoA dehydrogenase, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-04-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
8PFZ
 
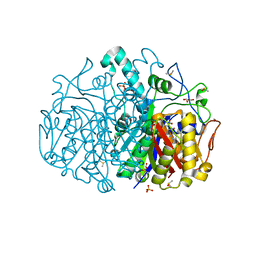 | | Pseudomonas aeruginosa FabF C164A mutant in complex with(S)-2-(1H-pyrazole-3-carboxamido)butanoic acid | | Descriptor: | (2~{S})-2-(1~{H}-pyrazol-3-ylcarbonylamino)butanoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2, CHLORIDE ION, ... | | Authors: | Georgiou, C, Brenk, R, Yadrykhinsky, V, Espeland, L.O. | | Deposit date: | 2023-06-17 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | New starting points for antibiotics targeting P. aeruginosa FabF discovered by crystallographic fragment screening followed by hit expansion
Chemrxiv, 2023
|
|
8PD1
 
 | | Pseudomonas aeruginosa FabF C164A mutant in complex with N-isopropyl-1H-imidazole-4-carboxamide | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Georgiou, C, Brenk, R, Yadrykhinsky, V, Espeland, L.O. | | Deposit date: | 2023-06-11 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | New starting points for antibiotics targeting P. aeruginosa FabF discovered by crystallographic fragment screening followed by hit expansion
Chemrxiv, 2023
|
|
6TIM
 
 | |
7O1K
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-E141A, beta-C92A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O4V
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with oxidized nicotinamide adenine dinucleotide | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-04-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
6WMP
 
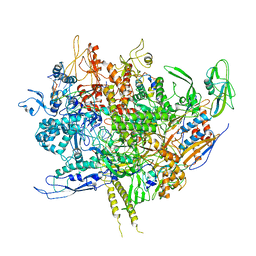 | | F. tularensis RNAPs70-iglA DNA complex | | Descriptor: | DNA NT-strand, DNA T-strand, DNA-directed RNA polymerase subunit alpha 1, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
8R1V
 
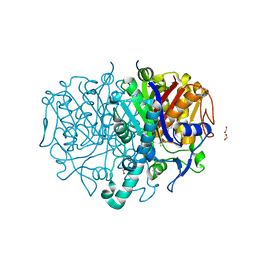 | | Pseudomonas aeruginosa FabF C164A in complex with N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)-2-(4-methoxyphenoxy)acetamide | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Yadrykhinsky, V, Brenk, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Design, quality and validation of the EU-OPENSCREEN fragment library poised to a high-throughput screening collection.
Rsc Med Chem, 15, 2024
|
|
8QM1
 
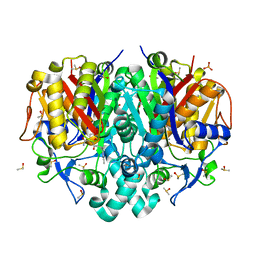 | | wild type Pa.FabF in complex cerulenin | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[acyl-carrier-protein] synthase 2, ... | | Authors: | Georgiou, C, Brenk, R, Espeland, L.O. | | Deposit date: | 2023-09-21 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | New starting points for antibiotics targeting P. aeruginosa FabF discovered by crystallographic fragment screening followed by hit expansion
Chemrxiv, 2023
|
|
8QER
 
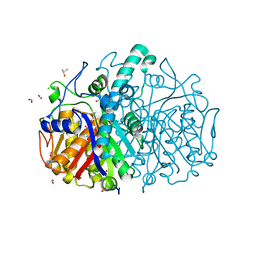 | | Pseudomonas aeruginosa FabF C164A in complex with 4-(1H-pyrazole-3-carbonylamino)pentanoic acid | | Descriptor: | (4~{R})-4-(1~{H}-pyrazol-3-ylcarbonylamino)pentanoic acid, 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] synthase 2, ... | | Authors: | Yadrykhins'ky, V, Brenk, R, Georgiou, C. | | Deposit date: | 2023-09-01 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New starting points for antibiotics targeting P. aeruginosa FabF discovered by crystallographic fragment screening followed by hit expansion
Chemrxiv, 2023
|
|
6YSV
 
 | |
8SSH
 
 | | MtrR from Neisseria gonorrhoeae bound to Ethinyl Estradiol | | Descriptor: | Ethinyl estradiol, HTH-type transcriptional regulator MtrR, PHOSPHATE ION | | Authors: | Hooks, G.M, Brennan, R.G. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Hormonal steroids induce multidrug resistance and stress response genes in Neisseria gonorrhoeae by binding to MtrR.
Nat Commun, 15, 2024
|
|
6OF0
 
 | | Structural basis for multidrug recognition and antimicrobial resistance by MTRR, an efflux pump regulator from Neisseria Gonorrhoeae | | Descriptor: | HTH-type transcriptional regulator MtrR, PHOSPHATE ION | | Authors: | Beggs, G.A, Kumaraswami, M, Shafer, W, Brennan, R.G. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Biochemical, and In Vivo Characterization of MtrR-Mediated Resistance to Innate Antimicrobials by the Human Pathogen Neisseria gonorrhoeae .
J.Bacteriol., 201, 2019
|
|
8PF8
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-72 | | Descriptor: | (2~{R})-3-bis[2-methyl-5-(trifluoromethyl)pyrazol-3-yl]boranyloxypropane-1,2-diol, 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystallographic fragment binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate channeling path between them
Biorxiv, 2024
|
|
7O1L
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-H462A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, COENZYME A, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O4U
 
 | |
7O1M
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-H462A, beta-C92A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7QH0
 
 | |
7O1I
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-E141A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, COENZYME A, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O1G
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-E141A-H462A, beta-C92A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, Putative acyltransferase Rv0859, SULFATE ION | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
6FZ5
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA and inhibitor bound | | Descriptor: | 4-[3-[(8~{a}~{R})-3,4,6,7,8,8~{a}-hexahydro-1~{H}-pyrrolo[1,2-a]pyrazin-2-yl]propyl]-2,6-bis(chloranyl)-~{N}-methyl-~{N}-(1,3,5-trimethylpyrazol-4-yl)benzenesulfonamide, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Kersten, F.C, Brenk, R. | | Deposit date: | 2018-03-14 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | How To Design Selective Ligands for Highly Conserved Binding Sites: A Case Study UsingN-Myristoyltransferases as a Model System.
J.Med.Chem., 63, 2020
|
|
6FZ3
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA and inhibitor bound | | Descriptor: | 2,6-bis(chloranyl)-4-[2-(4-methylpiperazin-1-yl)pyridin-4-yl]-~{N}-(1,3,5-trimethylpyrazol-4-yl)benzenesulfonamide, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Kersten, F.C, Brenk, R. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How To Design Selective Ligands for Highly Conserved Binding Sites: A Case Study UsingN-Myristoyltransferases as a Model System.
J.Med.Chem., 63, 2020
|
|
6TNM
 
 | | E. coli aerobic trifunctional enzyme subunit-alpha | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Fatty acid oxidation complex subunit alpha, GLYCEROL, ... | | Authors: | Sah-Teli, S.K, Hynonen, M.J, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2019-12-09 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Insights into the stability and substrate specificity of the E. coli aerobic beta-oxidation trifunctional enzyme complex.
J.Struct.Biol., 210, 2020
|
|
