2MFS
 
 | |
2K1V
 
 | | R3/I5 relaxin chimera | | Descriptor: | Insulin-like peptide INSL5, Relaxin-3 | | Authors: | Rosengren, K, Haugaard-Jonsson, L.M. | | Deposit date: | 2008-03-17 | | Release date: | 2008-04-08 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Structure of the R3/I5 Chimeric Relaxin Peptide, a Selective GPCR135 and GPCR142 Agonist.
J.Biol.Chem., 283, 2008
|
|
2LR9
 
 | |
2KNN
 
 | |
1Q71
 
 | | The structure of microcin J25 is a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone | | Descriptor: | microcin J25 | | Authors: | Rosengren, K.J, Clark, R, Daly, N.L, Goransson, U, Jones, A, Craik, D.J. | | Deposit date: | 2003-08-14 | | Release date: | 2003-12-16 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Microcin J25 has a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone.
J.Am.Chem.Soc., 125, 2003
|
|
2KNM
 
 | |
2MXM
 
 | |
2KBC
 
 | |
2KUX
 
 | |
1S7P
 
 | | Solution structure of thermolysin digested microcin J25 | | Descriptor: | microcin J25 | | Authors: | Rosengren, K.J, Blond, A, Afonso, C, Tabet, J.C, Rebuffat, S, Craik, D.J. | | Deposit date: | 2004-01-30 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of thermolysin cleaved microcin J25: extreme stability of a two-chain antimicrobial peptide devoid of covalent links
Biochemistry, 43, 2004
|
|
6A69
 
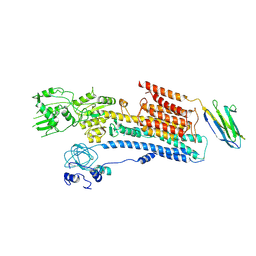 | | Cryo-EM structure of a P-type ATPase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroplastin, Plasma membrane calcium-transporting ATPase 1 | | Authors: | Gong, D.S, Chi, X.M, Ren, K, Huang, G.X.Y, Zhou, G.W, Yan, N, Lei, J.L, Zhou, Q. | | Deposit date: | 2018-06-27 | | Release date: | 2018-09-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Structure of the human plasma membrane Ca2+-ATPase 1 in complex with its obligatory subunit neuroplastin.
Nat Commun, 9, 2018
|
|
6AEG
 
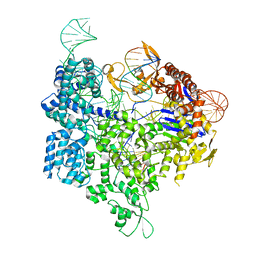 | | Crystal structure of xCas9 in complex with sgRNA and target DNA (GAT PAM) | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*AP*AP*GP*AP*TP*TP*AP*TP*TP*G)-3'), DNA nuclease, ... | | Authors: | Guo, M, Ren, K, Zhu, Y, Huang, Z. | | Deposit date: | 2018-08-04 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural insights into a high fidelity variant of SpCas9.
Cell Res., 29, 2019
|
|
6AEB
 
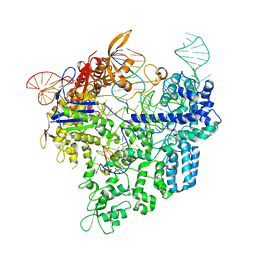 | | Crystal structure of xCas9 in complex with sgRNA and target DNA (AAG PAM) | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*AP*AP*AP*AP*GP*TP*AP*TP*TP*G)-3'), DNA Nuclease, ... | | Authors: | Guo, M, Ren, K, Zhu, Y, Huang, Z. | | Deposit date: | 2018-08-04 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structural insights into a high fidelity variant of SpCas9.
Cell Res., 29, 2019
|
|
5ID6
 
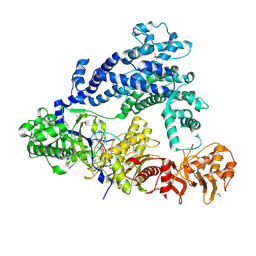 | | Structure of Cpf1/RNA Complex | | Descriptor: | Cpf1, MAGNESIUM ION, RNA (5'-R(P*AP*AP*UP*UP*UP*CP*UP*AP*CP*UP*AP*AP*GP*UP*GP*UP*AP*GP*AP*UP*C)-3') | | Authors: | Dong, D, Ren, K, Qiu, X, Wang, J, Huang, Z. | | Deposit date: | 2016-02-24 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | The crystal structure of Cpf1 in complex with CRISPR RNA
Nature, 532, 2016
|
|
6JGZ
 
 | | Structure of RyR2 (F/P/Ca2+ dataset) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, RyR2, ZINC ION | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-16 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JHN
 
 | | Structure of RyR2 (F/C/Ca2+ dataset) | | Descriptor: | CAFFEINE, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-18 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JH6
 
 | | Structure of RyR2 (F/A/Ca2+ dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Peptidyl-prolyl cis-trans isomerase FKBP1B, RyR2, ... | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JG3
 
 | | Cryo-EM structure of RyR2 (Ca2+ alone dataset) | | Descriptor: | Ryanodine receptor 2, ZINC ION | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-13 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
3ZOW
 
 | | Crystal Structure of Wild Type Nitrosomonas europaea Cytochrome c552 | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Karlsen, S, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-25 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
3ZOX
 
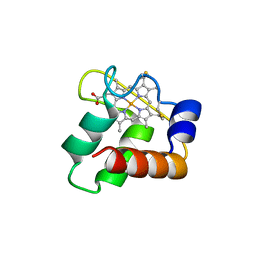 | | Crystal Structure of N64Del Mutant of Nitrosomonas europaea Cytochrome c552 (monoclinic space group) | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
6ENA
 
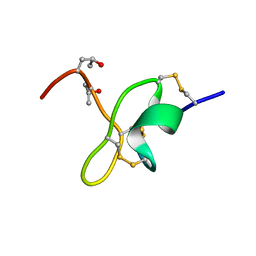 | | Nemertide alpha-1 | | Descriptor: | Nemertide alpha-1 | | Authors: | Jacobsson, E, Rosengren, K.J, Goransson, U. | | Deposit date: | 2017-10-04 | | Release date: | 2018-03-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Lineus longissimus, the longest animal on earth, expresses peptide toxins targeting voltage gated sodium channels
Sci Rep, 2018
|
|
7M29
 
 | |
7M28
 
 | |
7M3U
 
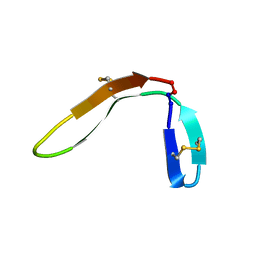 | |
