1YP5
 
 | | Yeast Myo5 SH3 domain, tetragonal crystal form | | Descriptor: | Myosin-5 isoform | | Authors: | Gonfloni, S, Kursula, P, Sacco, R, Cesareni, G, Wilmanns, M. | | Deposit date: | 2005-01-29 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Yeast Myo5 SH3 domain, tetragonal crystal form
To be Published
|
|
1B6Q
 
 | |
1XG2
 
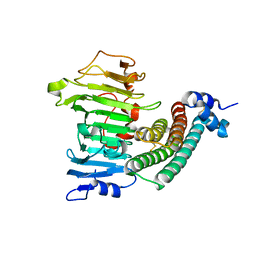 | | Crystal structure of the complex between pectin methylesterase and its inhibitor protein | | Descriptor: | Pectinesterase 1, Pectinesterase inhibitor | | Authors: | Di Matteo, A, Raiola, A, Camardella, L, Giovane, A, Bonivento, D, De Lorenzo, G, Cervone, F, Bellincampi, D, Tsernoglou, D. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Interaction between Pectin Methylesterase and a Specific Inhibitor Protein
Plant Cell, 17, 2005
|
|
1KL7
 
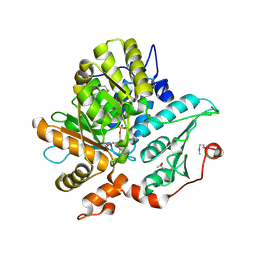 | | Crystal Structure of Threonine Synthase from Yeast | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Threonine Synthase | | Authors: | Garrido-Franco, M, Ehlert, S, Messerschmidt, A, Marinkovic, S, Huber, R, Laber, B, Bourenkov, G.P, Clausen, T. | | Deposit date: | 2001-12-11 | | Release date: | 2002-04-24 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of threonine synthase from yeast.
J.Biol.Chem., 277, 2002
|
|
1LBQ
 
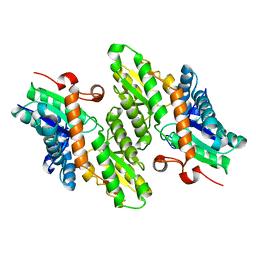 | | The crystal structure of Saccharomyces cerevisiae ferrochelatase | | Descriptor: | Ferrochelatase | | Authors: | Karlberg, T, Lecerof, D, Gora, M, Silvegren, G, Labbe-Bois, R, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-04-04 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal binding to Saccharomyces cerevisiae ferrochelatase
Biochemistry, 41, 2002
|
|
1LFW
 
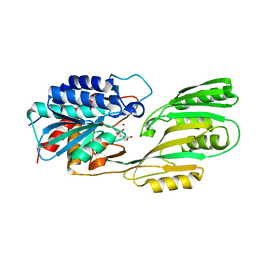 | | Crystal structure of pepV | | Descriptor: | 3-[(1-AMINO-2-CARBOXY-ETHYL)-HYDROXY-PHOSPHINOYL]-2-METHYL-PROPIONIC ACID, ZINC ION, pepV | | Authors: | Jozic, D, Bourenkow, G, Bartunik, H, Scholze, H, Dive, V, Henrich, B, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2002-04-12 | | Release date: | 2002-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Dinuclear Zinc Aminopeptidase PepV from Lactobacillus delbrueckii Unravels Its Preference for Dipeptides
Structure, 10, 2002
|
|
1AN2
 
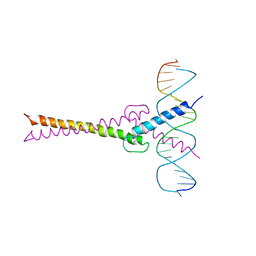 | | RECOGNITION BY MAX OF ITS COGNATE DNA THROUGH A DIMERIC B/HLH/Z DOMAIN | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*GP*GP*TP*CP*AP*CP*GP*TP*GP*AP*CP*C P*TP*AP*CP*AP*C)- 3'), PROTEIN (TRANSCRIPTION FACTOR MAX (TF MAX)) | | Authors: | Ferre-D'Amare, A.R, Prendergast, G.C, Ziff, E.B, Burley, S.K. | | Deposit date: | 1996-09-06 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain.
Nature, 363, 1993
|
|
1ZUY
 
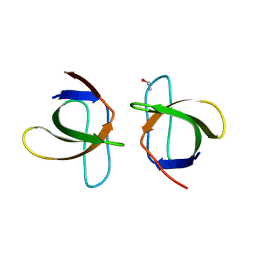 | | High-resolution structure of yeast Myo5 SH3 domain | | Descriptor: | ISOPROPYL ALCOHOL, Myosin-5 isoform | | Authors: | Wilmanns, M, Kursula, P, Gonfloni, S, Ferracuti, S, Cesareni, G. | | Deposit date: | 2005-06-01 | | Release date: | 2006-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | High-resolution structure of yeast Myo5 SH3 domain
To be published
|
|
1L8X
 
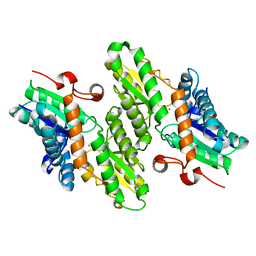 | | Crystal Structure of Ferrochelatase from the Yeast, Saccharomyces cerevisiae, with Cobalt(II) as the Substrate Ion | | Descriptor: | COBALT (II) ION, Ferrochelatase | | Authors: | Karlberg, T, Lecerof, D, Gora, M, Silvegren, G, Labbe-Bois, R, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-03-22 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Metal Binding to Saccharomyces cerevisiae Ferrochelatase
Biochemistry, 41, 2002
|
|
1KO1
 
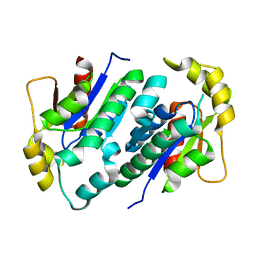 | | Crystal structure of gluconate kinase | | Descriptor: | CHLORIDE ION, Gluconate kinase | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
1Z48
 
 | | Crystal structure of reduced YqjM from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable NADH-dependent flavin oxidoreductase yqjM | | Authors: | Kitzing, K, Fitzpatrick, T.B, Wilken, C, Sawa, J, Bourenkov, G.P, Macheroux, P, Clausen, T. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.3 A Crystal Structure of the Flavoprotein YqjM Reveals a Novel Class of Old Yellow Enzymes
J.Biol.Chem., 280, 2005
|
|
1QXC
 
 | | NMR structure of the fragment 25-35 of beta amyloid peptide in 20/80 v:v hexafluoroisopropanol/water mixture | | Descriptor: | 11-mer peptide from Amyloid beta A4 protein | | Authors: | D'Ursi, A.M, Armenante, M.R, Guerrini, R, Salvadori, S, Sorrentino, G, Picone, D. | | Deposit date: | 2003-09-05 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide (25-35) in different media
J.Med.Chem., 47, 2004
|
|
1QWP
 
 | | NMR analysis of 25-35 fragment of beta amyloid peptide | | Descriptor: | 11-mer peptide from Amyloid beta A4 protein | | Authors: | D'Ursi, A.M, Armenante, M.R, Guerrini, R, Salvadori, S, Sorrentino, G, Picone, D. | | Deposit date: | 2003-09-03 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide (25-35) in different media
J.Med.Chem., 47, 2004
|
|
1QYT
 
 | | Solution structure of fragment (25-35) of beta amyloid peptide in SDS micellar solution | | Descriptor: | 11-mer peptide from Amyloid beta A4 protein | | Authors: | D'Ursi, A.M, Armenante, M.R, Guerrini, R, Salvadori, S, Sorrentino, G, Picone, D. | | Deposit date: | 2003-09-12 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide (25-35) in different media
J.Med.Chem., 47, 2004
|
|
1QX8
 
 | | Crystal structure of a five-residue deletion mutant of the Rop protein | | Descriptor: | Regulatory protein ROP | | Authors: | Glykos, N.M, Vlassi, M, Papanikolaou, Y, Kotsifaki, D, Cesareni, G, Kokkinidis, M. | | Deposit date: | 2003-09-04 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Loopless Rop: structure and dynamics of an engineered homotetrameric variant of the repressor of primer protein.
Biochemistry, 45, 2006
|
|
2AAJ
 
 | | Crystal Structures of the Wild-type, Mutant-P1A and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities | | Descriptor: | Malonate Semialdehyde Decarboxylase | | Authors: | Almrud, J.J, Poelarends, G.J, Johnson Jr, W.H, Serrano, H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2005-07-13 | | Release date: | 2005-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal Structures of the Wild-Type, P1A Mutant, and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities
Biochemistry, 44, 2005
|
|
2FIC
 
 | | The crystal structure of the BAR domain from human Bin1/Amphiphysin II and its implications for molecular recognition | | Descriptor: | Myc box-dependent-interacting protein 1, XENON | | Authors: | Casal, E, Federici, L, Zhang, W, Fernandez-Recio, J, Priego, E.M, Miguel, R.N, Duhadaway, J.B, Prendergast, G.C, Luisi, B.F, Laue, E.D. | | Deposit date: | 2005-12-29 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Crystal Structure of the BAR Domain from Human Bin1/Amphiphysin II and Its Implications for Molecular Recognition
Biochemistry, 45, 2006
|
|
1D7M
 
 | | COILED-COIL DIMERIZATION DOMAIN FROM CORTEXILLIN I | | Descriptor: | CORTEXILLIN I | | Authors: | Burkhard, P, Kammerer, R.A, Steinmetz, M.O, Bourenkov, G.P, Aebi, U. | | Deposit date: | 1999-10-19 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The coiled-coil trigger site of the rod domain of cortexillin I unveils a distinct network of interhelical and intrahelical salt bridges.
Structure Fold.Des., 8, 2000
|
|
2IYS
 
 | | Shikimate kinase from Mycobacterium tuberculosis in complex with shikimate, open LID (conf. A) | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, CHLORIDE ION, SHIKIMATE KINASE | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-21 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
2IYU
 
 | | Shikimate kinase from Mycobacterium tuberculosis in complex with ADP, open LID (conf. A) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, SHIKIMATE KINASE | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-22 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
2IYZ
 
 | | Shikimate kinase from Mycobacterium tuberculosis in complex with shikimate-3-phosphate and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SHIKIMATE KINASE, SHIKIMATE-3-PHOSPHATE | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-22 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
2IYR
 
 | | Shikimate kinase from Mycobacterium tuberculosis in complex with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, SHIKIMATE KINASE | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-21 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
2IYW
 
 | | Shikimate kinase from Mycobacterium tuberculosis in complex with MgATP, open LID (conf. B) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SHIKIMATE KINASE | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-22 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
2JAX
 
 | |
2IYT
 
 | | Shikimate kinase from Mycobacterium tuberculosis in unliganded state, open LID (conf. A) | | Descriptor: | CHLORIDE ION, SHIKIMATE KINASE | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-22 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
