2TMY
 
 | |
3SC2
 
 | | REFINED ATOMIC MODEL OF WHEAT SERINE CARBOXYPEPTIDASE II AT 2.2-ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SERINE CARBOXYPEPTIDASE II (CPDW-II), alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Liao, D.-I, Remington, S.J. | | Deposit date: | 1992-07-01 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined atomic model of wheat serine carboxypeptidase II at 2.2-A resolution.
Biochemistry, 31, 1992
|
|
2CSC
 
 | |
3RP2
 
 | | THE STRUCTURE OF RAT MAST CELL PROTEASE II AT 1.9-ANGSTROMS RESOLUTION | | Descriptor: | RAT MAST CELL PROTEASE II | | Authors: | Reynolds, R, Remington, S, Weaver, L, Fischer, R, Anderson, W, Ammon, H, Matthews, B. | | Deposit date: | 1984-09-10 | | Release date: | 1984-10-29 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of rat mast cell protease II at 1.9-A resolution.
Biochemistry, 27, 1988
|
|
3CSC
 
 | |
4EXO
 
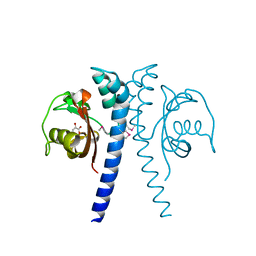 | | Revised, rerefined crystal structure of PDB entry 2QHK, methyl accepting chemotaxis protein | | Descriptor: | Methyl-accepting chemotaxis protein, PYRUVIC ACID | | Authors: | Sweeney, E.G, Henderson, J.N, Goers, J, Wreden, C, Hicks, K.G, Foster, J.K, Parthasarathy, R, Remington, S.J, Guillemin, K. | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Proposed Mechanism for the pH-Sensing Helicobacter pylori Chemoreceptor TlpB.
Structure, 20, 2012
|
|
3BLS
 
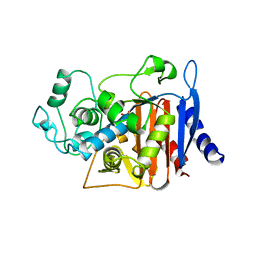 | | AMPC BETA-LACTAMASE FROM ESCHERICHIA COLI | | Descriptor: | AMPC BETA-LACTAMASE, M-AMINOPHENYLBORONIC ACID | | Authors: | Usher, K.C, Shoichet, B.K, Remington, S.J. | | Deposit date: | 1998-06-04 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of AmpC beta-lactamase from Escherichia coli bound to a transition-state analogue: possible implications for the oxyanion hypothesis and for inhibitor design.
Biochemistry, 37, 1998
|
|
2YFP
 
 | | STRUCTURE OF YELLOW-EMISSION VARIANT OF GFP | | Descriptor: | PROTEIN (GREEN FLUORESCENT PROTEIN) | | Authors: | Wachter, R.M, Elsliger, M.A, Kallio, K, Hanson, G.T, Remington, S.J. | | Deposit date: | 1998-08-17 | | Release date: | 1999-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of spectral shifts in the yellow-emission variants of green fluorescent protein.
Structure, 6, 1998
|
|
2H5Q
 
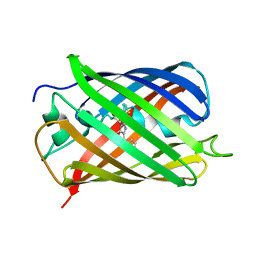 | | Crystal structure of mCherry | | Descriptor: | mCherry | | Authors: | Shu, X, Remington, S.J. | | Deposit date: | 2006-05-26 | | Release date: | 2006-08-22 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Novel Chromophores and Buried Charges Control Color in mFruits
Biochemistry, 45, 2006
|
|
2H5R
 
 | |
2H5P
 
 | |
2HQK
 
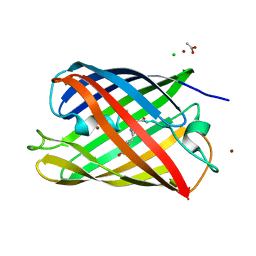 | | Crystal structure of a monomeric cyan fluorescent protein derived from Clavularia | | Descriptor: | ACETATE ION, CHLORIDE ION, Cyan fluorescent chromoprotein, ... | | Authors: | Henderson, J.N, Campbell, R.E, Ai, H, Remington, S.J. | | Deposit date: | 2006-07-18 | | Release date: | 2007-01-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging.
Biochem.J., 400, 2006
|
|
2H5O
 
 | | Crystal structure of mOrange | | Descriptor: | MAGNESIUM ION, mOrange | | Authors: | Shu, X, Remington, S.J. | | Deposit date: | 2006-05-26 | | Release date: | 2006-08-22 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Novel Chromophores and Buried Charges Control Color in mFruits
Biochemistry, 45, 2006
|
|
2QLI
 
 | | mPlum E16Q mutant | | Descriptor: | Fluorescent protein plum | | Authors: | Shu, X, Remington, S.J. | | Deposit date: | 2007-07-12 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural Studies of Far-Red Emission in mPlum, a Monomeric Red Fluorescent Protein
To be Published
|
|
3PSG
 
 | |
3TMY
 
 | | CHEY FROM THERMOTOGA MARITIMA (MN-III) | | Descriptor: | CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Usher, K.C, De La Cruz, A, Dahlquist, F.W, Remington, S.J. | | Deposit date: | 1997-06-04 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability.
Protein Sci., 7, 1998
|
|
3UB7
 
 | | Periplasmic portion of the Helicobacter pylori chemoreceptor TlpB with acetamide bound | | Descriptor: | ACETAMIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Henderson, J.N, Sweeney, E.G, Goers, J, Wreden, C, Hicks, K.G, Parthasarathy, R, Guillemin, K.J, Remington, S.J. | | Deposit date: | 2011-10-23 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB.
Structure, 20, 2012
|
|
3UB8
 
 | | Periplasmic portion of the Helicobacter pylori chemoreceptor TlpB with formamide bound | | Descriptor: | FORMAMIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Henderson, J.N, Sweeney, E.G, Goers, J, Wreden, C, Hicks, K.G, Parthasarathy, R, Guillemin, K.J, Remington, S.J. | | Deposit date: | 2011-10-23 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB.
Structure, 20, 2012
|
|
2GQ3
 
 | | mycobacterium tuberculosis malate synthase in complex with magnesium, malate, and coenzyme A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COENZYME A, D-MALATE, ... | | Authors: | Anstrom, D.M, Remington, S.J. | | Deposit date: | 2006-04-19 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The product complex of M. tuberculosis malate synthase revisited.
Protein Sci., 15, 2006
|
|
2DUI
 
 | |
2H8Q
 
 | |
2DUH
 
 | |
2DUE
 
 | |
2OTB
 
 | |
2OTE
 
 | | Crystal structure of a monomeric cyan fluorescent protein in the photobleached state | | Descriptor: | ACETATE ION, GFP-like fluorescent chromoprotein cFP484 | | Authors: | Henderson, J.N, Ai, H, Campbell, R.E, Remington, S.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for reversible photobleaching of a green fluorescent protein homologue.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
