2AQO
 
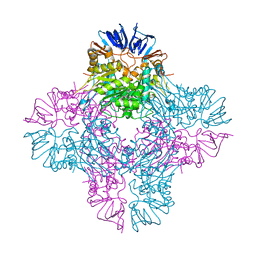 | | Crystal structure of E. coli Isoaspartyl Dipeptidase Mutant E77Q | | Descriptor: | Isoaspartyl dipeptidase, ZINC ION | | Authors: | Marti-Arbona, R, Thoden, J.B, Holden, H.M, Raushel, F.M. | | Deposit date: | 2005-08-18 | | Release date: | 2005-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional significance of Glu-77 and Tyr-137 within the active site of isoaspartyl dipeptidase.
Bioorg.Chem., 33, 2005
|
|
2Q08
 
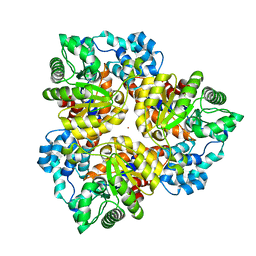 | | Crystal structure of the protein BH0493 from Bacillus halodurans C-125 complexed with ZN | | Descriptor: | BH0493 protein, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Patskovsky, Y, Sauder, J.M, Burley, S.K, Raushel, F, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-21 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Glucoronate Isomerase from Bacillus halodurans.
To be Published
|
|
6WN6
 
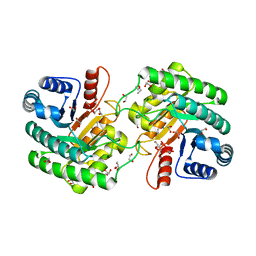 | | Crystal structure of 3-keto-D-glucoside 4-epimerase, YcjR, from E. coli, apo form | | Descriptor: | 1,2-ETHANEDIOL, 3-keto-D-glucoside 4-epimerase, MANGANESE (II) ION | | Authors: | Mabanglo, M.F, Raushel, F.M, Mukherjee, K. | | Deposit date: | 2020-04-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure and Reaction Mechanism of YcjR, an Epimerase That Facilitates the Interconversion of d-Gulosides to d-Glucosides inEscherichia coli.
Biochemistry, 59, 2020
|
|
2Q01
 
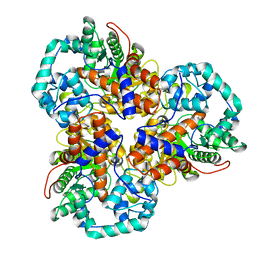 | | Crystal structure of glucuronate isomerase from Caulobacter crescentus | | Descriptor: | POTASSIUM ION, Uronate isomerase | | Authors: | Patskovsky, Y, Bonanno, J, Sridhar, V, Sauder, J.M, Freeman, J, Powell, A, Koss, J, Groshong, C, Gheyi, T, Wasserman, S.R, Raushel, F, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-18 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of Glucuronate Isomerase from Caulobacter crescentus.
To be Published
|
|
7M15
 
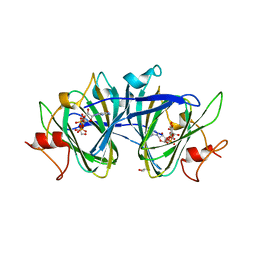 | | crystal structure of cj1430 in the presence of GDP-D-glycero-L-gluco-heptose, a GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase from campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, GDP-D-glycero-L-gluco-heptose, [(2R,3S,4R,5R)-5-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4R,5R,6S)-6-[(1R)-1,2-dihydroxyethyl]-3,4,5-trihydroxyoxan-2-yl dihydrogen diphosphate (non-preferred name) | | Authors: | Girardi, N.M, Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biosynthesis of d- glycero -l- gluco -Heptose in the Capsular Polysaccharides of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
8EWU
 
 | | X-ray structure of the GDP-6-deoxy-4-keto-D-lyxo-heptose-4-reductase from Campylobacter jejuni HS:15 | | Descriptor: | 1,2-ETHANEDIOL, GDP-L-fucose synthase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Xiang, D.F, Ghosh, M.K, Riegert, A.S, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bifunctional Epimerase/Reductase Enzymes Facilitate the Modulation of 6-Deoxy-Heptoses Found in the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 62, 2023
|
|
7M14
 
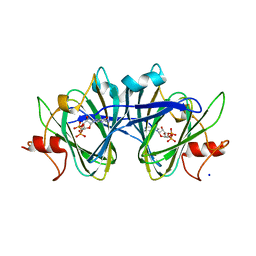 | | x-ray structure of cj1430 in the presence of GDP, a GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase from campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Girardi, N.M, Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biosynthesis of d- glycero -l- gluco -Heptose in the Capsular Polysaccharides of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
7M13
 
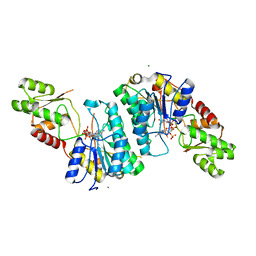 | | Crystal structure of CJ1428, a GDP-D-GLYCERO-L-GLUCO-HEPTOSE SYNTHASE from campylobacter jejuni in the presence of NADPH | | Descriptor: | 1,2-ETHANEDIOL, GDP-L-fucose synthase, MAGNESIUM ION, ... | | Authors: | Anderson, T.K, Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biosynthesis of d- glycero -l- gluco -Heptose in the Capsular Polysaccharides of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
1T36
 
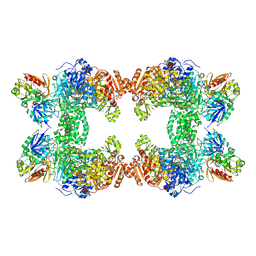 | | Crystal structure of E. coli carbamoyl phosphate synthetase small subunit mutant C248D complexed with uridine 5'-monophosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Carbamoyl-phosphate synthase large chain, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 2004-04-24 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Long-range allosteric transitions in carbamoyl phosphate synthetase.
Protein Sci., 13, 2004
|
|
7US5
 
 | | X-ray crystal structure of GDP-D-glycero-D-manno-heptose 4,6-Dehydratase from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, GDP-D-GLYCERO-D-MANNO-HEPTOSE 4,6-DEHYDRATASE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-04-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reaction Mechanism and Three-Dimensional Structure of GDP-d-glycero-alpha-d-manno-heptose 4,6-Dehydratase from Campylobacter jejuni.
Biochemistry, 61, 2022
|
|
7KWS
 
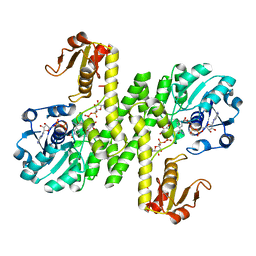 | | Cj1441 with NAD+ and UDP-glucose | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Riegert, A.S, Raushel, F.M. | | Deposit date: | 2020-12-02 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Functional and Structural Characterization of the UDP-Glucose Dehydrogenase Involved in Capsular Polysaccharide Biosynthesis from Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
2AQV
 
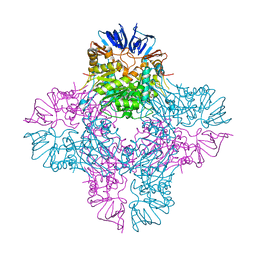 | | Crystal Structure of E. coli Isoaspartyl Dipeptidase mutant Y137F | | Descriptor: | Isoaspartyl dipeptidase, ZINC ION | | Authors: | Marti-Arbona, R, Thoden, J.B, Holden, H.M, Raushel, F.M. | | Deposit date: | 2005-08-18 | | Release date: | 2005-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional significance of Glu-77 and Tyr-137 within the active site of isoaspartyl dipeptidase.
Bioorg.Chem., 33, 2005
|
|
5HRM
 
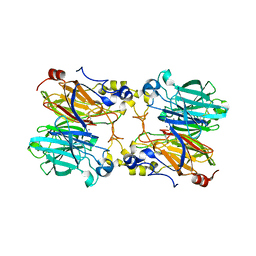 | |
6VO6
 
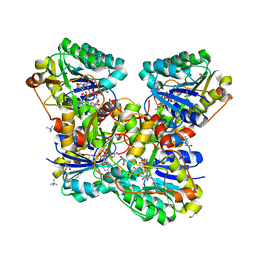 | | Crystal Structure of Cj1427, an Essential NAD-dependent Dehydrogenase from Campylobacter jejuni, in the Presence of NADH and GDP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Anderson, T.K, Spencer, K.D, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
6VO8
 
 | | X-ray structure of the Cj1427 in the presence of NADH and GDP-D-glycero-D-mannoheptose, an essential NAD-dependent dehydrogenase from Campylobacter jejuni | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Putative sugar-nucleotide epimerase/dehydratease, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{S},5~{S},6~{S})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Spencer, K.D, Anderson, T.K, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
3BE7
 
 | | Crystal structure of Zn-dependent arginine carboxypeptidase | | Descriptor: | ARGININE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Freeman, J, Iizuka, M, Bain, K, Rodgers, L, Raushel, F, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-16 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional identification of incorrectly annotated prolidases from the amidohydrolase superfamily of enzymes.
Biochemistry, 48, 2009
|
|
4ZSU
 
 | |
4ZST
 
 | |
5IOJ
 
 | |
8V4G
 
 | | X-ray structure of the NADP-dependent reductase from Campylobacter jejuni responsible for the synthesis of CDP-glucitol in the presence of CDP and NADP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, ... | | Authors: | Schumann, M.E, Thoden, J.B, Holden, H.M, Raushel, F.M. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of Cytidine Diphosphate-6-d-Glucitol for the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 63, 2024
|
|
8V4H
 
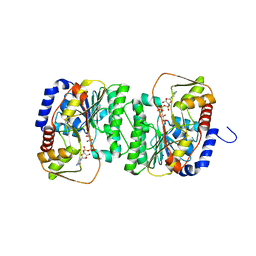 | | X-ray structure of the NADP-dependent reductase from Campylobacter jejuni responsible for the synthesis of CDP-glucitol in the presence of CDP-glucitol | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Putative nucleotide sugar dehydratase, ... | | Authors: | Thoden, J.B, Schumann, M.E, Holden, H.M, Raushel, F.M. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biosynthesis of Cytidine Diphosphate-6-d-Glucitol for the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 63, 2024
|
|
6P3K
 
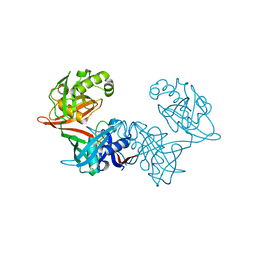 | | Crystal structure of LigU(C100S) | | Descriptor: | (4E)-oxalomesaconate Delta-isomerase, CHLORIDE ION | | Authors: | Cory, S.A, Hogancamp, T.N, Raushel, F.M, Barondeau, D.P. | | Deposit date: | 2019-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and Chemical Reaction Mechanism of LigU, an Enzyme That Catalyzes an Allylic Isomerization in the Bacterial Degradation of Lignin.
Biochemistry, 58, 2019
|
|
6P3H
 
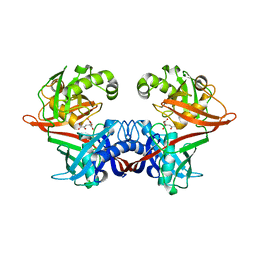 | | Crystal structure of LigU(K66M) bound to substrate | | Descriptor: | (1E)-4-oxobut-1-ene-1,2,4-tricarboxylic acid, (4E)-oxalomesaconate Delta-isomerase, CHLORIDE ION | | Authors: | Cory, S.A, Hogancamp, T.N, Raushel, F.M, Barondeau, D.P. | | Deposit date: | 2019-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and Chemical Reaction Mechanism of LigU, an Enzyme That Catalyzes an Allylic Isomerization in the Bacterial Degradation of Lignin.
Biochemistry, 58, 2019
|
|
6P3J
 
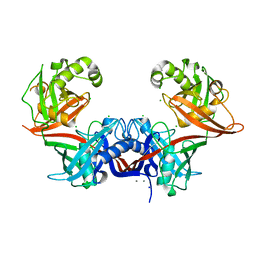 | | Crystal structure of LigU | | Descriptor: | (4E)-oxalomesaconate Delta-isomerase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cory, S.A, Hogancamp, T.N, Raushel, F.M, Barondeau, D.P. | | Deposit date: | 2019-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure and Chemical Reaction Mechanism of LigU, an Enzyme That Catalyzes an Allylic Isomerization in the Bacterial Degradation of Lignin.
Biochemistry, 58, 2019
|
|
3DUG
 
 | | Crystal structure of zn-dependent arginine carboxypeptidase complexed with zinc | | Descriptor: | ARGININE, GLYCEROL, ZINC ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Freeman, J, Iizuka, M, Bain, K, Rodgers, L, Raushel, F, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-17 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Functional identification of incorrectly annotated prolidases from the amidohydrolase superfamily of enzymes.
Biochemistry, 48, 2009
|
|
