1EDM
 
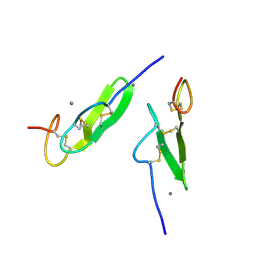 | | EPIDERMAL GROWTH FACTOR-LIKE DOMAIN FROM HUMAN FACTOR IX | | Descriptor: | CALCIUM ION, FACTOR IX | | Authors: | Rao, Z, Handford, P, Mayhew, M, Knott, V, Brownlee, G.G, Stuart, D. | | Deposit date: | 1996-03-21 | | Release date: | 1996-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of a Ca(2+)-binding epidermal growth factor-like domain: its role in protein-protein interactions.
Cell(Cambridge,Mass.), 82, 1995
|
|
2FRZ
 
 | |
2GR6
 
 | |
1GBS
 
 | |
2ESW
 
 | |
2FR7
 
 | | Crystal Structure of Cytochrome P450 CYP199A2 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, putative cytochrome P450 | | Authors: | Rao, Z, Wong, L.L, Xu, F, Bell, S.G. | | Deposit date: | 2006-01-19 | | Release date: | 2007-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of CYP199A2, a para-substituted benzoic acid oxidizing cytochrome P450 from Rhodopseudomonas palustris
J.Mol.Biol., 383, 2008
|
|
2GQX
 
 | | Crystal structure of cytochrome p450cam mutant (f87w/y96f/l244a/v247l/c334a) with pentachlorobenzene | | Descriptor: | 1,2,3,4,5-PENTACHLOROBENZENE, Cytochrome P450-cam, POTASSIUM ION, ... | | Authors: | Rao, Z, Wong, L.L, Xu, F, Bell, S.G. | | Deposit date: | 2006-04-22 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity correlations in pentachlorobenzene oxidation by engineered cytochrome P450cam
Protein Eng.Des.Sel., 20, 2007
|
|
1X0V
 
 | |
1X0X
 
 | |
1C78
 
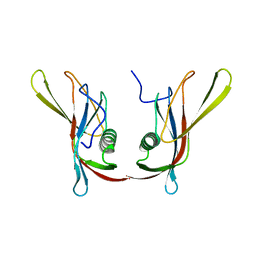 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
1C77
 
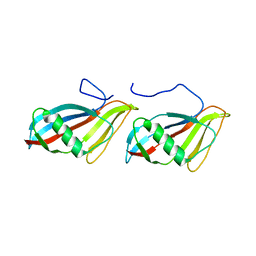 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
1C79
 
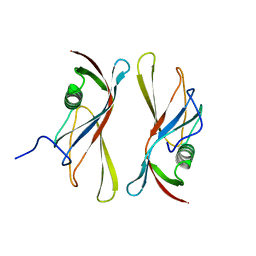 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
1C76
 
 | | STAPHYLOKINASE (SAK) MONOMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Staphylokinase Dimer Offers New Clue to Reduction of Immunogenicity
To be published
|
|
1ECW
 
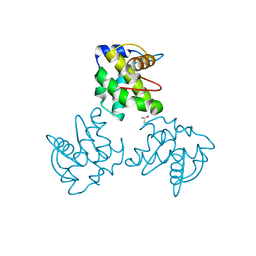 | | CRYSTAL STRUCTURE OF SIMIAN IMMUNODEFICIENCY VIRUS MATRIX ANTIGEN (SIV MA) AT 293K. | | Descriptor: | GAG POLYPROTEIN, ISOPROPYL ALCOHOL | | Authors: | Rao, Z, Belyaev, A, Fry, E, Roy, P, Jones, I.M, Stuart, D.I. | | Deposit date: | 2000-01-26 | | Release date: | 2000-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SIV matrix antigen and implications for virus assembly.
Nature, 378, 1995
|
|
1ED1
 
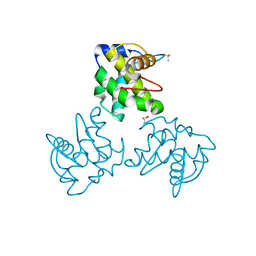 | | CRYSTAL STRUCTURE OF SIMIAN IMMUNODEFICIENCY VIRUS MATRIX ANTIGEN (SIV MA) AT 100K. | | Descriptor: | GAG POLYPROTEIN, ISOPROPYL ALCOHOL | | Authors: | Rao, Z, Belyaev, A, Fry, E, Roy, P, Jones, I.M, Stuart, D.I. | | Deposit date: | 2000-01-26 | | Release date: | 2000-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of SIV matrix antigen and implications for virus assembly.
Nature, 378, 1995
|
|
5XLL
 
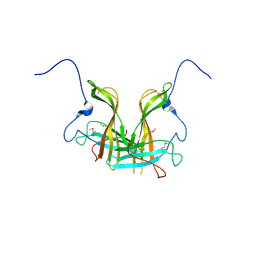 | | Dimer form of M. tuberculosis PknI sensor domain | | Descriptor: | Serine/threonine-protein kinase PknI | | Authors: | Rao, Z, Yan, Q. | | Deposit date: | 2017-05-10 | | Release date: | 2018-05-16 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural Insight into the Activation of PknI Kinase from M. tuberculosis via Dimerization of the Extracellular Sensor Domain.
Structure, 25, 2017
|
|
5XLM
 
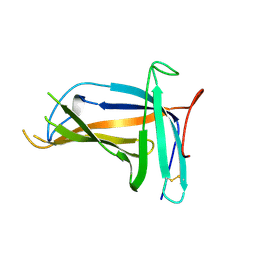 | | Monomer form of M.tuberculosis PknI sensor domain | | Descriptor: | Serine/threonine-protein kinase PknI | | Authors: | Rao, Z, Yan, Q. | | Deposit date: | 2017-05-10 | | Release date: | 2018-05-16 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into the Activation of PknI Kinase from M. tuberculosis via Dimerization of the Extracellular Sensor Domain.
Structure, 25, 2017
|
|
6KY3
 
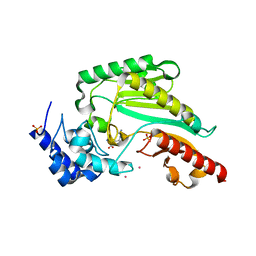 | | Structure of arginine kinase H284A mutant | | Descriptor: | ARGININE, Arginine kinase, PHOSPHATE ION, ... | | Authors: | Rao, Z, Park, J.H, Kim, S.Y, Kim, D.S. | | Deposit date: | 2019-09-16 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Insight into Structural Aspects of Histidine 284 of Daphnia magna Arginine Kinase.
Mol.Cells, 43, 2020
|
|
3CTM
 
 | | Crystal Structure of a Carbonyl Reductase from Candida Parapsilosis with anti-Prelog Stereo-specificity | | Descriptor: | Carbonyl Reductase | | Authors: | Zhang, R, Zhu, G, Li, X, Xu, Y, Zhang, X.C, Rao, Z. | | Deposit date: | 2008-04-14 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of a carbonyl reductase from Candida parapsilosis with anti-Prelog stereospecificity.
Protein Sci., 17, 2008
|
|
1MPW
 
 | | Molecular Recognition in (+)-a-Pinene Oxidation by Cytochrome P450cam | | Descriptor: | (+)-alpha-Pinene, CYTOCHROME P450CAM, POTASSIUM ION, ... | | Authors: | Bell, S.G, Chen, X, Sowden, R.J, Xu, F, Willams, J.N, Wong, L.-L, Rao, Z. | | Deposit date: | 2002-09-13 | | Release date: | 2002-10-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Molecular recognition in (+)-alpha-pinene oxidation by cytochrome P450cam
J.Am.Chem.Soc., 125, 2003
|
|
1N1A
 
 | | Crystal Structure of the N-terminal domain of human FKBP52 | | Descriptor: | FKBP52 | | Authors: | Li, P, Ding, Y, Wu, B, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2002-10-16 | | Release date: | 2002-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the N-terminal domain of human FKBP52.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5WTH
 
 | | Cryo-EM structure for Hepatitis A virus complexed with FAB | | Descriptor: | FAB Heavy Chain, FAB Light Chain, Polyprotein, ... | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-25 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTF
 
 | | Cryo-EM structure for Hepatitis A virus empty particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3VBH
 
 | | Crystal structure of formaldehyde treated human enterovirus 71 (space group R32) | | Descriptor: | CHLORIDE ION, Genome Polyprotein, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VBR
 
 | | Crystal structure of formaldehyde treated empty human Enterovirus 71 particle (room temperature) | | Descriptor: | Genome Polyprotein, capsid protein VP0, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
