8HIU
 
 | |
4QKQ
 
 | |
7BCW
 
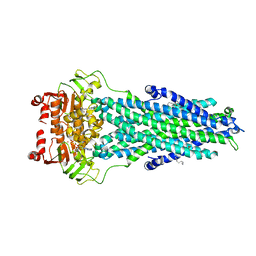 | | Structure of MsbA in Salipro with ADP vanadate | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-DIPHOSPHATE, ATP-dependent lipid A-core flippase, ... | | Authors: | Traore, D.A.K, Tidow, H. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of MsbA in saposin-lipid nanoparticles (Salipro) provides insights into nucleotide coordination.
Febs J., 289, 2022
|
|
6O1Z
 
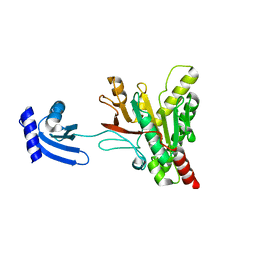 | | Structure of pCW3 conjugation coupling protein TcpA hexagonal crystal form | | Descriptor: | DNA translocase coupling protein | | Authors: | Traore, D.A.K, Ahktar, N, Torres, V.T, Adams, V, Coulibaly, F, Panjikar, S, Caradoc-Davies, T.T, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2019-02-22 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of pCW3 conjugation coupling protein TcpA
To be published
|
|
5VFY
 
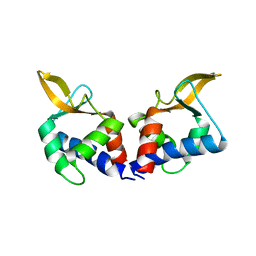 | | Structure of an accessory protein of the pCW3 relaxosome | | Descriptor: | TcpK | | Authors: | Traore, D.A.K, Wisniewski, J.A, Flanigan, S.F, Conroy, P.J, Panjikar, S, Mok, Y.-F, Lao, C, Griffin, M.D.W, Adams, V, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of TcpK in complex with oriT DNA of the antibiotic resistance plasmid pCW3.
Nat Commun, 9, 2018
|
|
5VFX
 
 | | Structure of an accessory protein of the pCW3 relaxosome in complex with the origin of transfer (oriT) DNA | | Descriptor: | TcpK, oriT | | Authors: | Traore, D.A.K, Wisniewski, J.A, Flanigan, S.F, Conroy, P.J, Panjikar, S, Mok, Y.-F, Lao, C, Griffin, M.D.W, Adams, V, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of TcpK in complex with oriT DNA of the antibiotic resistance plasmid pCW3.
Nat Commun, 9, 2018
|
|
6O1W
 
 | | Structure of pCW3 conjugation coupling protein TcpA monomer orthorhombic crystal form | | Descriptor: | DNA translocase coupling protein, beta-D-glucopyranose | | Authors: | Traore, D.A.K, Ahktar, N, Torres, V.T, Adams, V, Coulibaly, F, Panjikar, S, Caradoc-Davies, T.T, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2019-02-22 | | Release date: | 2020-02-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of pCW3 conjugation coupling protein TcpA
To be published
|
|
6O1Y
 
 | | Structure of pCW3 conjugation coupling protein TcpA monomeric form with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA translocase coupling protein, beta-D-glucopyranose | | Authors: | Traore, D.A.K, Ahktar, N, Torres, V.T, Adams, V, Coulibaly, F, Panjikar, S, Caradoc-Davies, T.T, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2019-02-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of pCW3 conjugation coupling protein TcpA
To be published
|
|
6O1X
 
 | | Structure of pCW3 conjugation coupling protein TcpA monomer form with ATPgS | | Descriptor: | DNA translocase coupling protein, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, beta-D-glucopyranose | | Authors: | Traore, D.A.K, Ahktar, N, Torres, V.T, Adams, V, Coulibaly, F, Panjikar, S, Caradoc-Davies, T.T, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2019-02-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure of pCW3 conjugation coupling protein TcpA
To be published
|
|
2FE3
 
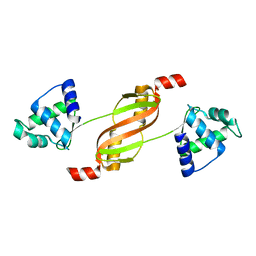 | |
3F8N
 
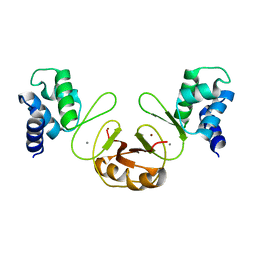 | | Crystal structure of PerR-Zn-Mn | | Descriptor: | MANGANESE (II) ION, Peroxide operon regulator, ZINC ION | | Authors: | Traore, D.A.K, Ferrer, J.-L, Jacquamet, L, Duarte, V, Latour, J.-M. | | Deposit date: | 2008-11-13 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding
Mol.Microbiol., 73, 2009
|
|
3W56
 
 | | Structure of a C2 domain | | Descriptor: | C2 domain protein | | Authors: | Traore, D.A.K, Whisstock, J.C. | | Deposit date: | 2013-01-24 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Defining the interaction of perforin with calcium and the phospholipid membrane.
Biochem.J., 456, 2013
|
|
3VKE
 
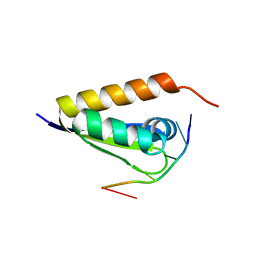 | |
2RGV
 
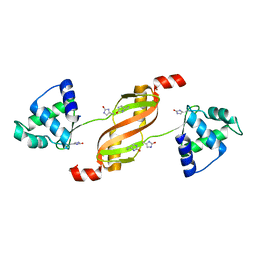 | |
3W57
 
 | | Structure of a C2 domain | | Descriptor: | C2 domain protein, CALCIUM ION | | Authors: | Traore, D.A.K, Whisstock, J.C. | | Deposit date: | 2013-01-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Defining the interaction of perforin with calcium and the phospholipid membrane.
Biochem.J., 456, 2013
|
|
3MIW
 
 | | Crystal Structure of Rotavirus NSP4 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural glycoprotein 4 | | Authors: | Chacko, A.R, Read, R.J, Dodson, E.J, Rao, D.C, Suguna, K. | | Deposit date: | 2010-04-12 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new pentameric structure of rotavirus NSP4 revealed by molecular replacement.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3N3I
 
 | | Crystal Structure of G48V/C95F tethered HIV-1 Protease/Saquinavir complex | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease | | Authors: | Prashar, V, Bihani, S.C, Das, A, Rao, D.R, Hosur, M.V. | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Insights into the mechanism of drug resistance: X-ray structure analysis of G48V/C95F tethered HIV-1 protease dimer/saquinavir complex
Biochem.Biophys.Res.Commun., 396, 2010
|
|
6ACA
 
 | | Crystal structure of Mycobacterium tuberculosis Mfd at 3.6 A resolution | | Descriptor: | Mycobacterium tuberculosis Mfd | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-26 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
2AOQ
 
 | | Crystal structure of MutH-unmethylated DNA complex | | Descriptor: | 5'-D(*GP*CP*AP*TP*GP*AP*TP*CP*AP*TP*GP*C)-3', CALCIUM ION, DNA mismatch repair protein mutH | | Authors: | Lee, J.Y, Chang, J, Joseph, N, Ghirlando, R, Rao, D.N, Yang, W. | | Deposit date: | 2005-08-13 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage.
Mol.Cell, 20, 2005
|
|
6AC6
 
 | | Ab initio crystal structure of Selenomethionine labelled Mycobacterium smegmatis Mfd | | Descriptor: | Mycobacterium smegmatis Mfd, SULFATE ION | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6AC8
 
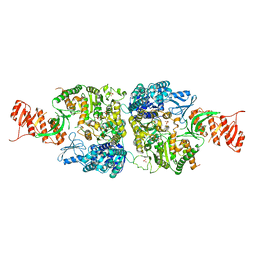 | | Crystal structure of Mycobacterium smegmatis Mfd at 2.75 A resolution | | Descriptor: | Mycobacterium smegmatis Mfd, SULFATE ION | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6ACX
 
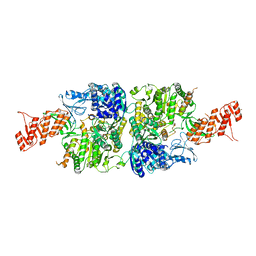 | | Crystal structure of Mycobacterium smegmatis Mfd in complex with ADP + Pi at 3.5 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Mycobacterium smegmatis Mfd, PHOSPHATE ION, ... | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
2AOR
 
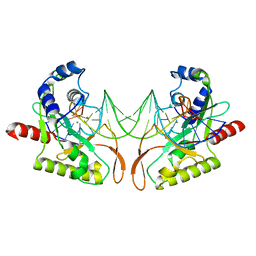 | | Crystal structure of MutH-hemimethylated DNA complex | | Descriptor: | 5'-D(*CP*AP*GP*GP*(6MA)P*TP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*AP*TP*CP*CP*TP*G)-3', CALCIUM ION, DNA mismatch repair protein mutH | | Authors: | Lee, J.Y, Chang, J, Joseph, N, Ghirlando, R, Rao, D.N, Yang, W. | | Deposit date: | 2005-08-13 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage.
Mol.Cell, 20, 2005
|
|
4DTF
 
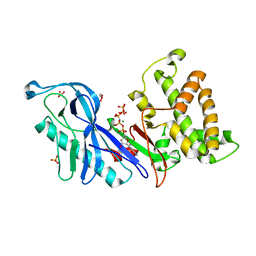 | | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with AMP-PNP and Mg++ | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-02-21 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with AMP-PNP and Mg++
J.Biol.Chem., 2012
|
|
2OSE
 
 | | Crystal Structure of the Mimivirus Cyclophilin | | Descriptor: | CHLORIDE ION, Probable peptidyl-prolyl cis-trans isomerase | | Authors: | Eisenmesser, E.Z, Thai, V, Renesto, P, Raoult, D. | | Deposit date: | 2007-02-05 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural, biochemical, and in vivo characterization of the first virally encoded cyclophilin from the Mimivirus.
J.Mol.Biol., 378, 2008
|
|
