1A80
 
 | | Native 2,5-DIKETO-D-GLUCONIC acid reductase a from CORYNBACTERIUM SP. complexed with nadph | | Descriptor: | 2,5-DIKETO-D-GLUCONIC ACID REDUCTASE A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Khurana, S, Powers, D.B, Anderson, S, Blaber, M. | | Deposit date: | 1998-03-31 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of 2,5-diketo-D-gluconic acid reductase A complexed with NADPH at 2.1-A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4RRO
 
 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | Descriptor: | (4S,4a'R,10a'S)-2-amino-8'-(2-fluoropyridin-3-yl)-1,4a'-dimethyl-3',4',4a',10a'-tetrahydro-2'H-spiro[imidazole-4,10'-pyrano[3,2-b]chromen]-5(1H)-one, Beta-secretase 1, NICKEL (II) ION | | Authors: | Thomas, A.A, Hunt, K.W, Newhouse, B, Watts, R.J, Liu, X, Vigers, G.P.A, Smith, D, Rhodes, S.P, Brown, K.D, Otten, J.N, Burkard, M, Cox, A.A, Geck Do, M.K, Dutcher, D, Rana, S, DeLisle, R.K, Regal, K, Wright, A.D, Groneberg, R, Liao, J, Scearce-Levie, K, Siu, M, Purkey, H.E, Lyssikatos, J.P. | | Deposit date: | 2014-11-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
4RRS
 
 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | Descriptor: | (4R,4a'R,10a'S)-8'-(2-fluoropyridin-3-yl)-4a'-methyl-3',4',4a',10a'-tetrahydro-2'H-spiro[1,3-oxazole-4,10'-pyrano[3,2-b]chromen]-2-amine, Beta-secretase 1, NICKEL (II) ION | | Authors: | Thomas, A.A, Hunt, K.W, Newhouse, B, Watts, R.J, Liu, X, Vigers, G.P.A, Smith, D, Rhodes, S.P, Brown, K.D, Otten, J.N, Burkard, M, Cox, A.A, Geck Do, M.K, Dutcher, D, Rana, S, DeLisle, R.K, Regal, K, Wright, A.D, Groneberg, R, Liao, J, Scearce-Levie, K, Siu, M, Purkey, H.E, Lyssikatos, J.P. | | Deposit date: | 2014-11-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
4RRN
 
 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | Descriptor: | (4S,4a'S,10a'R)-2-amino-8'-(2-fluoropyridin-3-yl)-1-methyl-3',4',4a',10a'-tetrahydro-2'H-spiro[imidazole-4,10'-pyrano[3,2-b]chromen]-5(1H)-one, Beta-secretase 1, NICKEL (II) ION | | Authors: | Thomas, A.A, Hunt, K.W, Newhouse, B, Watts, R.J, Liu, X, Vigers, G.P.A, Smith, D, Rhodes, S.P, Brown, K.D, Otten, J.N, Burkard, M, Cox, A.A, Geck Do, M.K, Dutcher, D, Rana, S, DeLisle, R.K, Regal, K, Wright, A.D, Groneberg, R, Liao, J, Scearce-Levie, K, Siu, M, Purkey, H.E, Lyssikatos, J.P. | | Deposit date: | 2014-11-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
4PP7
 
 | | Highly Potent and Selective 3-N-methylquinazoline-4(3H)-one Based Inhibitors of B-RafV600E Kinase | | Descriptor: | N-{2,4-difluoro-3-[methyl(3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]phenyl}propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Wenglowsky, S, Ren, L, Grina, J, Hansen, J.D, Laird, E.R, Moreno, D, Dinkel, V, Gloor, S.L, Hastings, G, Rana, S, Rasor, K, Sturgis, H.L, Voegtli, W.C, Vigers, G.P.A, Willis, B, Mathieu, S, Rudolph, J. | | Deposit date: | 2014-02-26 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Highly potent and selective 3-N-methylquinazoline-4(3H)-one based inhibitors of B-Raf(V600E) kinase.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2WLS
 
 | | Crystal structure of Mus musculus Acetylcholinesterase in complex with AMTS13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pang, Y.P, Ekstrom, F, Polsinelli, G.A, Gao, Y, Rana, S, Hua, D.H, Andersson, B, Andersson, P.O, Peng, L, Singh, S.K, Mishra, R.K, Zhu, K.Y, Fallon, A.M, Ragsdale, D.W, Brimijoin, S. | | Deposit date: | 2009-06-25 | | Release date: | 2009-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective and Irreversible Inhibitors of Mosquito Acetylcholinesterases for Controlling Malaria and Other Mosquito-Borne Diseases.
Plos One, 4, 2009
|
|
2FBD
 
 | | The crystallographic structure of the digestive lysozyme 1 from Musca domestica at 1.90 Ang. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lysozyme 1, SULFATE ION | | Authors: | Cancado, F.C, Marana, S.R, Barbosa, J.A.R.G. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallization, data collection and phasing of two digestive lysozymes from Musca domestica.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
5CG0
 
 | | Crystal structure of Spodoptera frugiperda Beta-glycosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tamaki, F.K, Souza, D.P, Souza, V.P, Farah, S.C, Marana, S.R. | | Deposit date: | 2015-07-08 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Using the Amino Acid Network to Modulate the Hydrolytic Activity of beta-Glycosidases.
PLoS ONE, 11, 2016
|
|
8UA7
 
 | | Medusavirus Nucleosome Core Particle | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Toner, C.M, Hoitsma, N.M, Weerawarana, S, Luger, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Characterization of Medusavirus encoded histones reveals nucleosome-like structures and a unique linker histone.
Nat Commun, 15, 2024
|
|
6VDG
 
 | | Crystal Structure of the Y182A HisF Mutant from Thermotoga maritima | | Descriptor: | Imidazole glycerol phosphate synthase subunit HisF, PHOSPHATE ION | | Authors: | Almeida, V.M, Matsuyama, B.Y, Farah, C.S, Marana, S.R. | | Deposit date: | 2019-12-27 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Role of a high centrality residue in protein dynamics and thermal stability.
J.Struct.Biol., 213, 2021
|
|
2HUG
 
 | | 3D Solution Structure of the Chromo-2 Domain of cpSRP43 complexed with cpSRP54 peptide | | Descriptor: | Signal recognition particle 43 kDa protein, chloroplast, Signal recognition particle 54 kDa protein | | Authors: | Kathir, K.M, Vaithiyalingam, S, Henry, R, Thallapuranam, S.K.K. | | Deposit date: | 2006-07-26 | | Release date: | 2007-09-18 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Assembly of chloroplast signal recognition particle involves structural rearrangement in cpSRP43.
J.Mol.Biol., 381, 2008
|
|
1RVH
 
 | | SOLUTION STRUCTURE OF THE DNA DODECAMER GCAAAATTTTGC | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*AP*TP*TP*TP*TP*GP*C)-3' | | Authors: | Stefl, R, Wu, H, Ravindranathan, S, Sklenar, V, Feigon, J. | | Deposit date: | 2003-12-13 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | DNA A-tract bending in three dimensions: Solving the dA4T4 vs. dT4A4 conundrum.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2H5Z
 
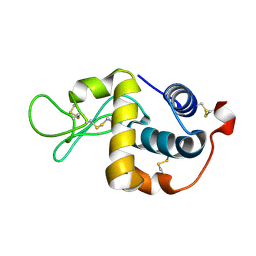 | | Crystallographic structure of digestive lysozyme 1 from Musca domestica bound to chitotetraose at 1.92 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme 1 | | Authors: | Valerio, A.A, Cancado, F.C, Marana, S.R, Barbosa, J.A.R.G. | | Deposit date: | 2006-05-29 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystallization, data collection and phasing of two digestive lysozymes from Musca domestica
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1RVI
 
 | | SOLUTION STRUCTURE OF THE DNA DODECAMER CGTTTTAAAACG | | Descriptor: | 5'-D(*CP*GP*TP*TP*TP*TP*AP*AP*AP*AP*CP*G)-3' | | Authors: | Stefl, R, Wu, H, Ravindranathan, S, Sklenar, V, Feigon, J. | | Deposit date: | 2003-12-13 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | DNA A-tract bending in three dimensions: Solving the dA4T4 vs. dT4A4 conundrum.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3CAF
 
 | |
3F05
 
 | |
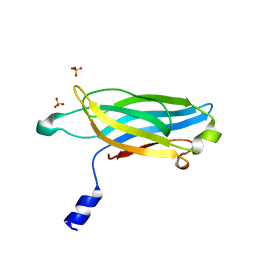
3F04
 
 | |
3CU1
 
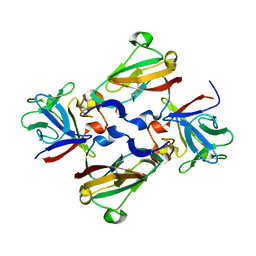 | | Crystal Structure of 2:2:2 FGFR2D2:FGF1:SOS complex | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Fibroblast growth factor receptor 2, Heparin-binding growth factor 1 | | Authors: | Guo, F, Dakshinamurthy, R, Thallapuranam, S.K.K, Sakon, J. | | Deposit date: | 2008-04-15 | | Release date: | 2009-04-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of 2:2:2 FGFR2D2:FGF1:SOS complex
To be Published
|
|
3F01
 
 | |
3EUU
 
 | |
3CB7
 
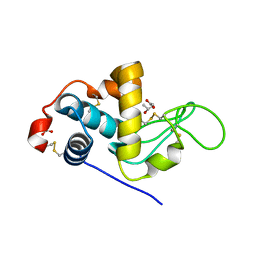 | | The crystallographic structure of the digestive lysozyme 2 from Musca domestica at 1.9 Ang. | | Descriptor: | ACETIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Cancado, F.C, Valerio, A.A, Marana, S.R, Barbosa, J.A.R.G. | | Deposit date: | 2008-02-21 | | Release date: | 2009-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystallographic structure of the digestive lysozyme 2 from Musca domestica at 1.9 Ang.
To be Published
|
|
3F00
 
 | |
2OKS
 
 | | X-ray Structure of a DNA Repair Substrate Containing an Alkyl Interstrand Crosslink at 1.65 Resolution | | Descriptor: | 5'-D(*CP*CP*AP*AP*(C34)P*GP*TP*TP*GP*G)-3', CALCIUM ION | | Authors: | Swenson, M.C, Paranawithana, S.R, Miller, P.S, Kielkopf, C.L. | | Deposit date: | 2007-01-17 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of a DNA repair substrate containing an alkyl interstrand cross-link at 1.65 a resolution.
Biochemistry, 46, 2007
|
|
2N3O
 
 | | Structure of PTB RRM1(41-163) bound to an RNA stemloop containing a structured loop derived from viral internal ribosomal entry site RNA | | Descriptor: | Polypyrimidine tract-binding protein 1, RNA (5'-R(*GP*GP*GP*AP*CP*CP*UP*GP*GP*UP*CP*UP*UP*UP*CP*CP*AP*GP*GP*UP*CP*CP*C)-3') | | Authors: | Maris, C, Jayne, S.F, Damberger, F.F, Ravindranathan, S, Allain, F.H.-T. | | Deposit date: | 2015-06-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | C-terminal helix folding upon pyrimidine-rich hairpin binding to PTB RRM1. Implications for PTB function in Encephalomyocarditis virus IRES activity.
To be Published
|
|
1XCB
 
 | | X-ray Structure of a Rex-Family Repressor/NADH Complex from Thermus Aquaticus | | Descriptor: | CALCIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Redox-sensing transcriptional repressor rex | | Authors: | Sickmier, E.A, Brekasis, D, Paranawithana, S, Bonanno, J.B, Burley, S.K, Paget, M.S, Kielkopf, C.L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-09-01 | | Release date: | 2004-09-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-Ray Structure of a Rex-Family Repressor/NADH Complex: Insights into the Mechanism of Redox Sensing
Structure, 13, 2005
|
|
