6QZU
 
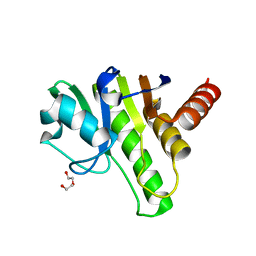 | | Getah virus macro domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Non-structural polyprotein | | Authors: | Ferreira Ramos, A.S, Sulzenbacher, G, Coutard, B. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6R0F
 
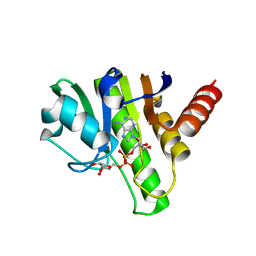 | | Getah virus macro domain in complex with ADPr, pose 1 | | Descriptor: | Non-structural polyprotein, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Sulzenbacher, G, Ferreira Ramos, A.S, Coutard, B. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6R0R
 
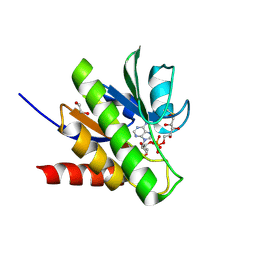 | | Getah virus macro domain in complex with ADPr covalently bond to Cys34 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural polyprotein, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{S})-2,3,4,5-tetrakis(oxidanyl)pentyl] hydrogen phosphate | | Authors: | Ferreira Ramos, A.S, Sulzenbacher, G, Coutard, B. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
2OPV
 
 | |
6SAI
 
 | |
2KXH
 
 | | Solution structure of the first two RRM domains of FIR in the complex with FBP Nbox peptide | | Descriptor: | Poly(U)-binding-splicing factor PUF60, peptide of Far upstream element-binding protein 1 | | Authors: | Cukier, C.D, Ramos, A, Hollingworth, D, Diaz-Moreno, I, Kelly, G. | | Deposit date: | 2010-05-05 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of FIR-mediated c-myc transcriptional control.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6QEY
 
 | | IMP1 KH1 and KH2 domains create a structural platform with unique RNA recognition and re-modelling properties | | Descriptor: | ACETONITRILE, Insulin-like growth factor 2 mRNA-binding protein 1, PHOSPHATE ION | | Authors: | Dagil, R, Ball, N.J, Ogrodowicz, R.W, Purkiss, A.G, Taylor, I.A, Ramos, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | IMP1 KH1 and KH2 domains create a structural platform with unique RNA recognition and re-modelling properties.
Nucleic Acids Res., 47, 2019
|
|
2KXF
 
 | | Solution structure of the first two RRM domains of FBP-interacting repressor (FIR) | | Descriptor: | Poly(U)-binding-splicing factor PUF60 | | Authors: | Cukier, C.D, Ramos, A, Hollingworth, D, Diaz-Moreno, I, Kelly, G. | | Deposit date: | 2010-05-04 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of FIR-mediated c-myc transcriptional control.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2X1A
 
 | | Structure of Rna15 RRM with RNA bound (G) | | Descriptor: | 5'-R(*GP*UP*UP*GP*UP)-3', MAGNESIUM ION, MRNA 3'-END-PROCESSING PROTEIN RNA15 | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2X1F
 
 | | Structure of Rna15 RRM with bound RNA (GU) | | Descriptor: | 5'-R(*GP*UP*UP*GP*UP)-3', MRNA 3'-END-PROCESSING PROTEIN RNA15 | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2X1B
 
 | | Structure of RNA15 RRM | | Descriptor: | MRNA 3'-END-PROCESSING PROTEIN RNA15, PHOSPHATE ION | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2JQP
 
 | | NMR structure determination of Bungatoxin from Bungarus candidus (Malayan Krait) | | Descriptor: | Weak toxin 1 | | Authors: | Vivekanandan, S, Jois, S.D, Kini, R.M, Troncone, L.R.P, de Magalhaes, L, Ujikawa, G.Y, Ramos, A.T. | | Deposit date: | 2007-06-06 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution Structure of Bungatoxin from Bungarus Candidus (Malayan Krait)venom
To be Published
|
|
