8CTO
 
 | |
8CUN
 
 | |
8CWA
 
 | |
2RN7
 
 | | NMR solution structure of TnpE protein from Shigella flexneri. Northeast Structural Genomics Target SfR125 | | 分子名称: | IS629 orfA | | 著者 | Ramelot, T.A, Cort, J.R, Semesi, A, Garcia, M, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-12-08 | | 公開日 | 2008-01-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of TnpE protein from Shigella flexneri. Northeast
Structural Genomics Target SfR125
To be Published
|
|
7M5T
 
 | |
1JJG
 
 | | Solution Structure of Myxoma Virus Protein M156R | | 分子名称: | M156R | | 著者 | Ramelot, T.A, Cort, J.R, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2001-07-05 | | 公開日 | 2002-03-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Myxoma virus immunomodulatory protein M156R is a structural mimic of eukaryotic translation initiation factor eIF2alpha.
J.Mol.Biol., 322, 2002
|
|
7TZD
 
 | |
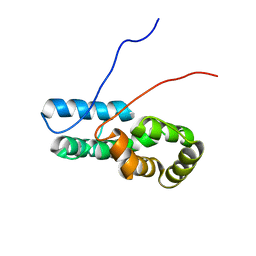
7TZ8
 
 | |
7UBF
 
 | |
7UBD
 
 | |
7UBI
 
 | |
7UZL
 
 | |
7UBE
 
 | |
7UBG
 
 | |
7UBC
 
 | |
7UBH
 
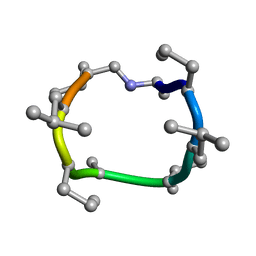 | |
2B3W
 
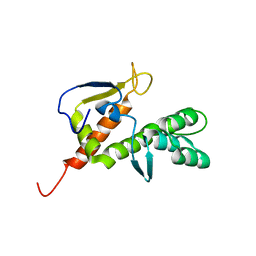 | | NMR structure of the E.coli protein YbiA, Northeast Structural Genomics target ET24. | | 分子名称: | Hypothetical protein ybiA | | 著者 | Ramelot, T.A, Cort, J.R, Xiao, R, Shih, L.Y, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2005-09-21 | | 公開日 | 2005-11-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the E.coli protein YbiA, Northeast Structural Genomics target ET24.
To be Published
|
|
2DSM
 
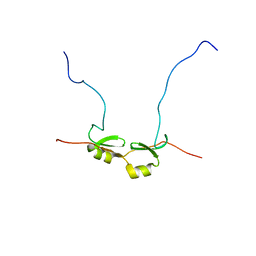 | | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450 | | 分子名称: | Hypothetical protein yqaI | | 著者 | Ramelot, T.A, Cort, J.R, Wang, D, Janua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-07-01 | | 公開日 | 2006-08-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450
to be published
|
|
1XPW
 
 | | Solution NMR Structure of human protein HSPCO34. Northeast Structural Genomics Target HR1958 | | 分子名称: | LOC51668 protein | | 著者 | Ramelot, T.A, Xiao, R, Ma, L.C, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2004-10-09 | | 公開日 | 2004-11-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study.
Proteins, 75, 2009
|
|
1YWU
 
 | | Solution NMR structure of Pseudomonas Aeruginosa protein PA4608. Northeast Structural Genomics target PaT7 | | 分子名称: | hypothetical protein PA4608 | | 著者 | Ramelot, T.A, Yee, A.A, Cort, J.R, Semesi, A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2005-02-18 | | 公開日 | 2005-03-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein.
Proteins, 66, 2007
|
|
1M94
 
 | | Solution Structure of the Yeast Ubiquitin-Like Modifier Protein Hub1 | | 分子名称: | Protein YNR032c-a | | 著者 | Ramelot, T.A, Cort, J.R, Yee, A.A, Semesi, A, Edwards, A.M, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2002-07-26 | | 公開日 | 2002-12-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Yeast Ubiquitin-Like Modifier Protein Hub1
J.STRUCT.FUNCT.GENOM., 4, 2003
|
|
1NXI
 
 | | Solution structure of Vibrio cholerae protein VC0424 | | 分子名称: | conserved hypothetical protein VC0424 | | 著者 | Ramelot, T.A, Ni, S, Goldsmith-Fischman, S, Cort, J.R, Honig, B, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2003-02-10 | | 公開日 | 2003-07-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Vibrio cholerae protein VC0424: a variation of the ferredoxin-like fold.
Protein Sci., 12, 2003
|
|
1Q48
 
 | | Solution NMR Structure of The Haemophilus Influenzae Iron-Sulfur Cluster Assembly Protein U (IscU) with Zinc Bound at the Active Site. Northeast Structural Genomics Consortium Target IR24. This protein is not apo, it is a model without zinc binding constraints. | | 分子名称: | NifU-like protein | | 著者 | Ramelot, T.A, Cort, J.R, Xiao, R, Shastry, R, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2003-08-01 | | 公開日 | 2003-11-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site.
J.Mol.Biol., 344, 2004
|
|
1LV3
 
 | | Solution NMR Structure of Zinc Finger Protein yacG from Escherichia coli. Northeast Structural Genomics Consortium Target ET92. | | 分子名称: | HYPOTHETICAL PROTEIN YacG, ZINC ION | | 著者 | Ramelot, T.A, Cort, J.R, Yee, A.A, Semesi, A, Edwards, A.M, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2002-05-24 | | 公開日 | 2002-09-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the Escherichia coli protein YacG: a novel sequence motif in the zinc-finger family of proteins.
Proteins, 49, 2002
|
|
1R9P
 
 | | Solution NMR Structure Of The Haemophilus Influenzae Iron-Sulfur Cluster Assembly Protein U (IscU) with Zinc Bound at the Active Site. Northeast Structural Genomics Consortium Target IR24. | | 分子名称: | NifU-like protein, ZINC ION | | 著者 | Ramelot, T.A, Cort, J.R, Xiao, R, Shastry, R, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2003-10-30 | | 公開日 | 2004-11-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site.
J.Mol.Biol., 344, 2004
|
|
