8P3L
 
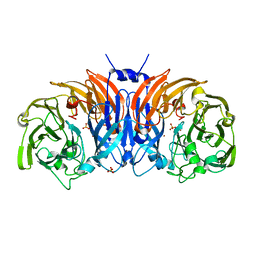 | | The structure of thiocyanate dehydrogenase mutant form with Thr 169 replaced by Ala from Thioalkalivibrio paradoxus | | Descriptor: | COPPER (II) ION, SULFATE ION, Twin-arginine translocation signal domain-containing protein | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Komolov, A.S, Rakitina, T.V, Dergousova, N.I, Dorovatovskii, P.V, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-05-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Improvement of the Diffraction Properties of Thiocyanate Dehydrogenase Crystals
Crystallography Reports, 2023
|
|
8P3M
 
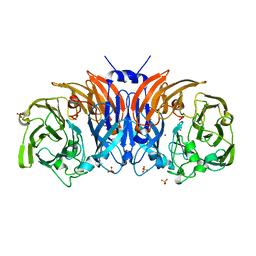 | | The structure of thiocyanate dehydrogenase mutant form with Lys 281 replaced by Ala from Thioalkalivibrio paradoxus | | Descriptor: | BORIC ACID, COPPER (II) ION, SODIUM ION, ... | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Komolov, A.S, Rakitina, T.V, Dergousova, N.I, Dorovatovskii, P.V, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-05-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Improvement of the Diffraction Properties of Thiocyanate Dehydrogenase Crystals
Crystallography Reports, 2023
|
|
6ERK
 
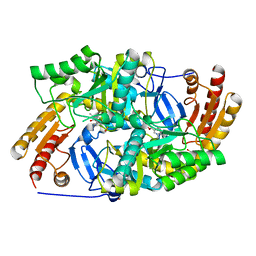 | | Crystal structure of diaminopelargonic acid aminotransferase from Psychrobacter cryohalolentis | | Descriptor: | 1,2-ETHANEDIOL, Aminotransferase, GLYCEROL, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bezsudnova, E.Y, Stekhanova, T.N, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2017-10-18 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diaminopelargonic acid transaminase from Psychrobacter cryohalolentis is active towards (S)-(-)-1-phenylethylamine, aldehydes and alpha-diketones.
Appl. Microbiol. Biotechnol., 102, 2018
|
|
6Q8E
 
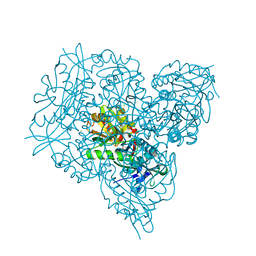 | | Crystal structure of branched-chain amino acid aminotransferase from Thermobaculum terrenum in PMP-form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Branched-chain-amino-acid aminotransferase, CHLORIDE ION | | Authors: | Boyko, K.M, Bezsudnova, E.Y, Nikolaeva, A.Y, Zeifman, Y.S, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-12-14 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural insights into PLP fold type IV transaminase from Thermobaculum terrenum.
Biochimie, 158, 2018
|
|
7Z79
 
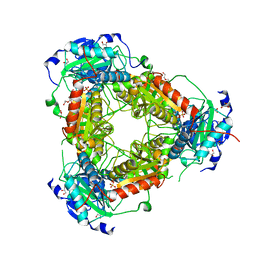 | | Crystal structure of aminotransferase-like protein from Variovorax paradoxus | | Descriptor: | Aminotransferase, class 4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boyko, K.M, Matyuta, I.O, Nikolaeva, A.Y, Khrenova, M.G, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Puzzling Protein from Variovorax paradoxus Has a PLP Fold Type IV Transaminase Structure and Binds PLP without Catalytic Lysine
Crystals, 12, 2022
|
|
6UWE
 
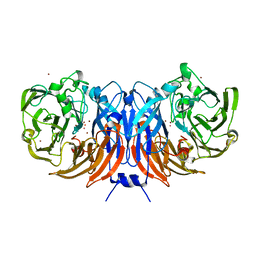 | | Crystal structure of recombinant thiocyanate dehydrogenase from Thioalkalivibrio paradoxus saturated with copper | | Descriptor: | COPPER (II) ION, UNKNOWN ATOM OR ION, thiocyanate dehydrogenase | | Authors: | Shabalin, I.G, Osipov, E, Tikhonova, T.V, Rakitina, T.V, Boyko, K.M, Popov, V.O. | | Deposit date: | 2019-11-05 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7O9U
 
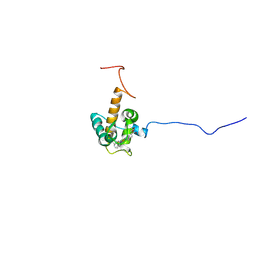 | | Solution structure of oxidized cytochrome c552 from Thioalkalivibrio paradoxus | | Descriptor: | Cytochrome c552, HEME C | | Authors: | Britikov, V.V, Britikova, E.V, Altukhov, D.A, Timofeev, V.I, Dergousova, N.I, Rakitina, T.V, Tikhonova, T.V, Usanov, S.A, Popov, V.O, Bocharov, E.V. | | Deposit date: | 2021-04-17 | | Release date: | 2021-05-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Unusual Cytochrome c 552 from Thioalkalivibrio paradoxus : Solution NMR Structure and Interaction with Thiocyanate Dehydrogenase.
Int J Mol Sci, 23, 2022
|
|
6SJI
 
 | | The structure of thiocyanate dehydrogenase from Thioalkalivibrio paradoxus mutant with His 482 replaced by Gln | | Descriptor: | COPPER (II) ION, SULFATE ION, thiocyanate dehydrogenase | | Authors: | Polyakov, K.M, Tikhonova, T.V, Rakitina, T.V, Osipov, E, Popov, V.O. | | Deposit date: | 2019-08-13 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4HGX
 
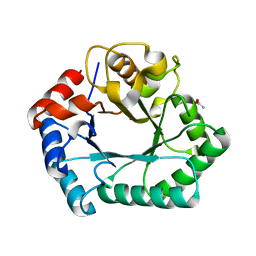 | | Crystal structure of xylose isomerase domain containing protein (stm4435) from salmonella typhimurium lt2 with unknown ligand | | Descriptor: | ACETATE ION, Xylose isomerase domain containing protein, ZINC ION | | Authors: | Boyko, K.M, Gorbacheva, M.A, Korzhenevskiy, D.A, Dorovatovsky, P.V, Rakitina, T.V, Lipkin, A.V, Shumilin, I.A, Minor, W, Popov, V.O. | | Deposit date: | 2012-10-09 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of xylose isomerase domain containing protein (stm4435) from salmonella typhimurium lt2 with unknown ligand
To be Published
|
|
9J0V
 
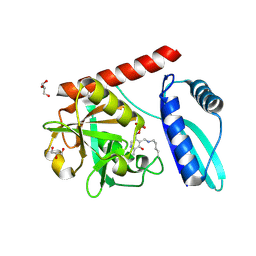 | | Crystal structure of monomeric PLP-dependent transaminase from Desulfobacula toluolica in P 21 21 21 space group | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dat: predicted D-alanine aminotransferase, GLYCEROL, ... | | Authors: | Matyuta, I.O, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-08-03 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | High/low resolution monomeric PLP-dependent transaminase from Desulfobacula toluolica
To Be Published
|
|
9J0U
 
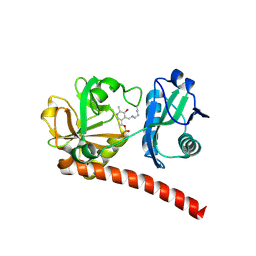 | | Crystal structure of monomeric PLP-dependent transaminase from Desulfobacula toluolica in F 41 3 2 space group | | Descriptor: | Dat: predicted D-alanine aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-08-03 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | High/low resolution monomeric PLP-dependent transaminase from Desulfobacula toluolica
To Be Published
|
|
6ZHK
 
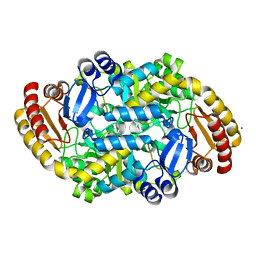 | | Crystal structure of adenosylmethionine-8-amino-7-oxononanoate aminotransferase from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, MAGNESIUM ION | | Authors: | Boyko, K.M, Nikolaeva, T.N, Stekhanova, T.N, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2020-06-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-Dimensional Structure of Thermostable D-Amino Acid Transaminase from the Archaeon Methanocaldococcus jannaschii DSM 2661
Crystallography Reports, 2021
|
|
5L8Z
 
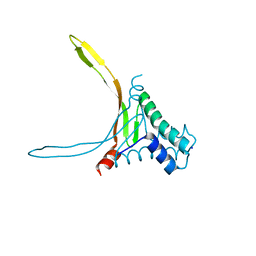 | | Structure of thermostable DNA-binding HU protein from micoplasma Spiroplasma melliferum | | Descriptor: | DNA-binding protein, SODIUM ION | | Authors: | Boyko, K.M, Gorbacheva, M.A, Rakitina, T.V, Korzhenevskiy, D.A, Kamashev, D.E, Vanyushkina, A.A, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the high thermal stability of the histone-like HU protein from the mollicute Spiroplasma melliferum KC3.
Sci Rep, 6, 2016
|
|
8BPN
 
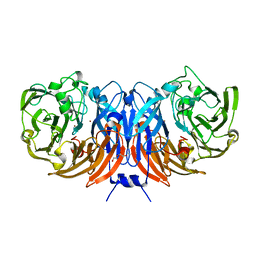 | | The structure of thiocyanate dehydrogenase mutant form with Phe 436 replaced by Gln from Thioalkalivibrio paradoxus | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, Twin-arginine translocation signal domain-containing protein | | Authors: | Varfolomeeva, L.A, Solovieva, A.Y, Shipkov, N.S, Kulikova, O.G, Dergousova, N.I, Rakitina, T.V, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-01-09 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Probing the Role of a Conserved Phenylalanine in the Active Site of Thiocyanate Dehydrogenase
Crystals, 12, 2022
|
|
4RGZ
 
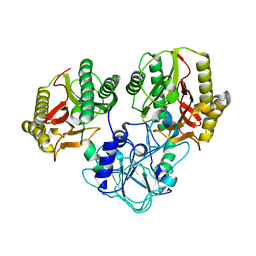 | | Crystal structure of recombinant prolidase from Thermococcus sibiricus at P21221 spacegroup | | Descriptor: | PHOSPHATE ION, Xaa-Pro aminopeptidase, ZINC ION | | Authors: | Timofeev, V.I, Korgenevsky, D.A, Gorbacheva, M.A, Boyko, K.M, Slutsky, E, Rakitina, T.V, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2014-10-01 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of recombinant prolidase from Thermococcus sibiricus in space group P21221.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8ONO
 
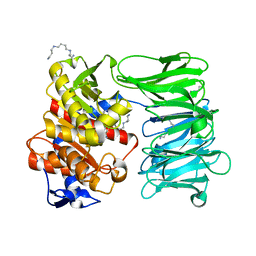 | | Modified oligopeptidase B from S. proteamaculans in intermediate conformation with 5 spermine molecule at 1.65 A resolution | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2023-04-03 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 5 spermine molecule at 1.65 A resolution
To Be Published
|
|
4WWV
 
 | | Aminopeptidase APDkam598 from the archaeon Desulfurococcus kamchatkensis | | Descriptor: | Aminopeptidase from family M42 | | Authors: | Petrova, T, Boyko, K.M, Rakitina, T.V, Korzhenevskiy, D.A, Gorbacheva, M.A, Popov, V.O. | | Deposit date: | 2014-11-12 | | Release date: | 2015-03-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.006 Å) | | Cite: | Structure of the dodecamer of the aminopeptidase APDkam598 from the archaeon Desulfurococcus kamchatkensis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
7P7X
 
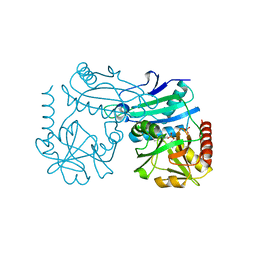 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis (holo form). | | Descriptor: | ACETATE ION, Aminotransferase class IV, PHOSPHATE ION, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bakunova, A.K, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2021-07-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Uncommon Active Site of D-Amino Acid Transaminase from Haliscomenobacter hydrossis : Biochemical and Structural Insights into the New Enzyme.
Molecules, 26, 2021
|
|
6GKR
 
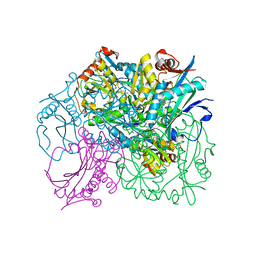 | | Crystal structure of branched-chain amino acid aminotransferase from Thermobaculum terrenum in PLP-form (holo-form) | | Descriptor: | ACETATE ION, Branched-chain-amino-acid aminotransferase, CHLORIDE ION, ... | | Authors: | Boyko, K.M, Bezsudnova, E.Y, Nikolaeva, A.Y, Zeifman, Y.S, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-05-21 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Biochemical and structural insights into PLP fold type IV transaminase from Thermobaculum terrenum.
Biochimie, 158, 2018
|
|
6H65
 
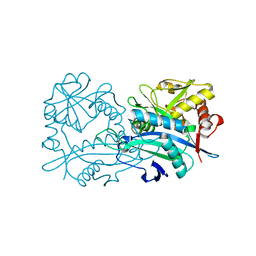 | | Crystal structure of the branched-chain-amino-acid aminotransferase from Haliangium ochraceum | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Timofeev, V.I, Bezsudnova, E.Y, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-07-26 | | Release date: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the branched-chain-amino-acid aminotransferase from Haliangium ochraceum
To Be Published
|
|
2NDP
 
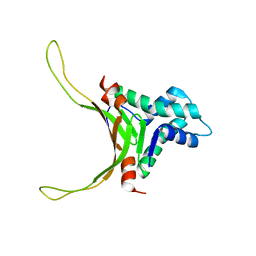 | | Structure of DNA-binding HU protein from micoplasma Mycoplasma gallisepticum | | Descriptor: | Histone-like DNA-binding superfamily protein | | Authors: | Altukhov, D.A, Talyzina, A.A, Agapova, Y.K, Vlaskina, A.V, Korzhenevskiy, D.A, Bocharov, E.V, Rakitina, T.V, Timofeev, V.I, Popov, V.O. | | Deposit date: | 2016-09-13 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Enhanced conformational flexibility of the histone-like (HU) protein from Mycoplasma gallisepticum.
J.Biomol.Struct.Dyn., 36, 2018
|
|
4NPA
 
 | | Scrystal structure of protein with unknown function from Vibrio cholerae at P22121 spacegroup | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Putative uncharacterized protein, ... | | Authors: | Boyko, K.M, Gorbacheva, M.A, Rakitina, T.V, Korgenevsky, D.A, Dorovatovsky, P.V, Lipkin, A.V, Minor, W, Shumilin, I.A, Popov, V.O. | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of protein with unknown function from Vibrio cholerae at P22121 spacegroup
To be Published
|
|
4NPO
 
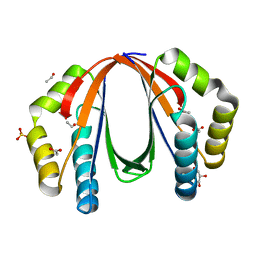 | | Crystal structure of protein with unknown function from Deinococcus radiodurans at P61 spacegroup | | Descriptor: | ACETATE ION, ACETYL GROUP, GLYCEROL, ... | | Authors: | Boyko, K.M, Gorbacheva, M.A, Rakitina, T.V, Korgenevsky, D.A, Shabalin, I.G, Shumilin, I.A, Dorovatovsky, P.V, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure of protein with unknown function at P61 spacegroup
To be Published
|
|
7NE7
 
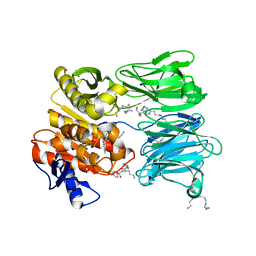 | | oligopeptidase B from S. proteomaculans with modified hinge region in complex with N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide | | Descriptor: | N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide, Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | Deposit date: | 2021-02-03 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of N alpha-p-tosyl-lysyl Chloromethylketone-Bound Oligopeptidase B from Serratia Proteamaculans Revealed a New Type of Inhibitor Binding
Crystals, 11, 2021
|
|
7NE4
 
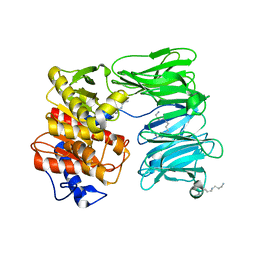 | | E125A mutant of oligopeptidase B from S. proteomaculans with modified hinge region | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | First Crystal Structure of Bacterial Oligopeptidase B in an Intermediate State: The Roles of the Hinge Region Modification and Spermine.
Biology (Basel), 10, 2021
|
|
