1JEF
 
 | | TURKEY LYSOZYME COMPLEX WITH (GLCNAC)3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME, SULFATE ION | | Authors: | Harata, K, Muraki, M. | | Deposit date: | 1997-04-23 | | Release date: | 1997-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | X-ray structure of turkey-egg lysozyme complex with tri-N-acetylchitotriose. Lack of binding ability at subsite A.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
5CAB
 
 | | Structure of Leishmania nucleoside diphostate kinase mutant Del5-Cterm | | Descriptor: | Nucleoside diphosphate kinase, SULFATE ION | | Authors: | Vieira, P.S, de Giuseppe, P.O, de Oliveira, A.H.C, Murakami, M.T. | | Deposit date: | 2015-06-29 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.953 Å) | | Cite: | The role of the C-terminus and Kpn loop in the quaternary structure stability of nucleoside diphosphate kinase from Leishmania parasites.
J.Struct.Biol., 192, 2015
|
|
5C7P
 
 | |
5CAA
 
 | |
3O5S
 
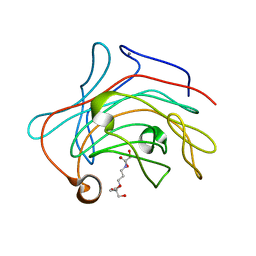 | | Crystal Structure of the endo-beta-1,3-1,4 glucanase from Bacillus subtilis (strain 168) | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION | | Authors: | Santos, C.R, Tonoli, C.C.C, Souza, A.R, Furtado, G.P, Ribeiro, L.F, Ward, R.J, Murakami, M.T. | | Deposit date: | 2010-07-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural characterization of a Beta-1,3 1,4-glucanase from Bacillus subtilis 168
PROCESS BIOCHEM, 46, 2011
|
|
1IQB
 
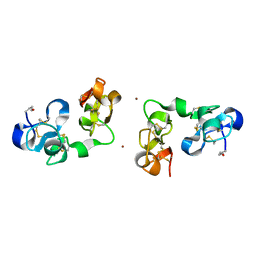 | |
4DCF
 
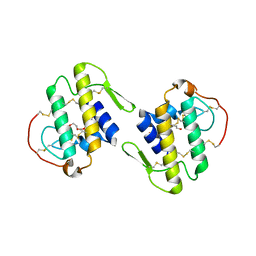 | | Structure of MTX-II from Bothrops brazili | | Descriptor: | MTX-II chain A, TETRAETHYLENE GLYCOL | | Authors: | Ullah, A, Souza, T.A.C.B, Betzel, C, Murakami, M.T, Arni, R.K. | | Deposit date: | 2012-01-17 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic portrayal of different conformational states of a Lys49 phospholipase A2 homologue: insights into structural determinants for myotoxicity and dimeric configuration.
Int.J.Biol.Macromol., 51, 2012
|
|
2RRI
 
 | | NMR structure of vasoactive intestinal peptide in DPC Micelle | | Descriptor: | Vasoactive intestinal peptide | | Authors: | Umetsu, Y, Tenno, T, Goda, N, Shirakawa, M, Ikegami, T, Hiroaki, H. | | Deposit date: | 2010-12-21 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural difference of vasoactive intestinal peptide in two distinct membrane-mimicking environments
Biochim.Biophys.Acta, 1814, 2011
|
|
2RQQ
 
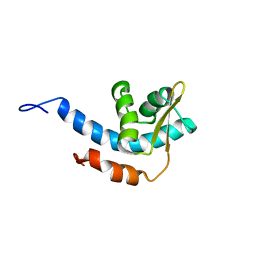 | | Structure of C-terminal region of Cdt1 | | Descriptor: | DNA replication factor Cdt1 | | Authors: | Jee, J.G, Mizuno, T, Kamada, K, Tochio, H, Hiroaki, H, Hanaoka, F, Shirakawa, M. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and mutagenesis studies of the C-terminal region of licensing factor Cdt1 enable the identification of key residues for binding to replicative helicase Mcm proteins
J.Biol.Chem., 285, 2010
|
|
2RPA
 
 | | The solution structure of N-terminal domain of microtubule severing enzyme | | Descriptor: | Katanin p60 ATPase-containing subunit A1 | | Authors: | Iwaya, N, Kuwahara, Y, Unzai, S, Nagata, T, Tomii, K, Goda, N, Tochio, H, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2008-05-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A common substrate recognition mode conserved between katanin P60 and VPS4 governs microtubule severing and membrane skeleton reorganization
J.Biol.Chem., 285, 2010
|
|
2MX2
 
 | | UBX-L domain of VCIP135 | | Descriptor: | Deubiquitinating protein VCIP135 | | Authors: | Iwazu, T, Murayama, S, Igarashi, R, Hrioaki, H, Shirakawa, M, Tochio, H. | | Deposit date: | 2014-12-07 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and interaction mode of the UBX-L domain of VCIP135 determined by solution NMR spectroscopy
To be Published
|
|
1FR0
 
 | | SOLUTION STRUCTURE OF THE HISTIDINE-CONTAINING PHOSPHOTRANSFER DOMAIN OF ANAEROBIC SENSOR KINASE ARCB FROM ESCHERICHIA COLI. | | Descriptor: | ARCB | | Authors: | Ikegami, T, Okada, T, Ohki, I, Hirayama, J, Mizuno, T, Shirakawa, M. | | Deposit date: | 2000-09-07 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamic character of the histidine-containing phosphotransfer domain of anaerobic sensor kinase ArcB from Escherichia coli.
Biochemistry, 40, 2001
|
|
1COP
 
 | |
1COO
 
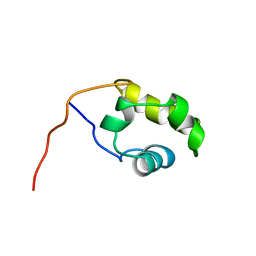 | | THE COOH-TERMINAL DOMAIN OF RNA POLYMERASE ALPHA SUBUNIT | | Descriptor: | RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Jeon, Y.H, Negishi, T, Shirakawa, M, Yamazaki, T, Fujita, N, Ishihama, A, Kyogoku, Y. | | Deposit date: | 1995-10-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the activator contact domain of the RNA polymerase alpha subunit.
Science, 270, 1995
|
|
1TBY
 
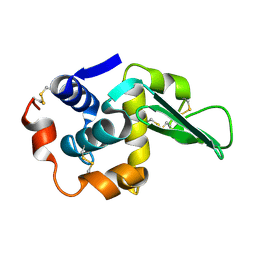 | |
2P3F
 
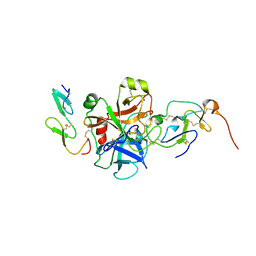 | | Crystal structure of the factor Xa/NAP5 complex | | Descriptor: | Anti-coagulant protein 5, Coagulation factor X, SODIUM ION | | Authors: | Rios-Steiner, J.L, Murakami, M.T, Tulinsky, A, Arni, R.K. | | Deposit date: | 2007-03-08 | | Release date: | 2007-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Active and exo-site inhibition of human factor Xa: structure of des-Gla factor Xa inhibited by NAP5, a potent nematode anticoagulant protein from Ancylostoma caninum
J.Mol.Biol., 371, 2007
|
|
1TAY
 
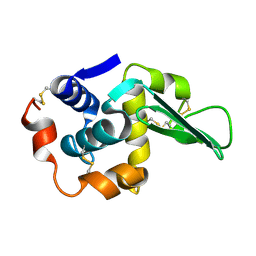 | |
1TDY
 
 | |
1TCY
 
 | |
1WLF
 
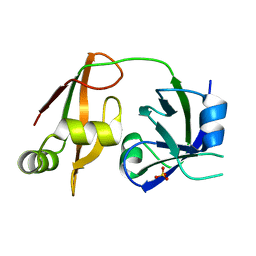 | | Structure of the N-terminal domain of PEX1 AAA-ATPase: Characterization of a putative adaptor-binding domain | | Descriptor: | Peroxisome biogenesis factor 1, SULFATE ION | | Authors: | Shiozawa, K, Maita, N, Tomii, K, Seto, A, Goda, N, Tochio, H, Akiyama, Y, Shimizu, T, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2004-06-25 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the N-terminal Domain of PEX1 AAA-ATPase: CHARACTERIZATION OF A PUTATIVE ADAPTOR-BINDING DOMAIN
J.Biol.Chem., 279, 2004
|
|
2D07
 
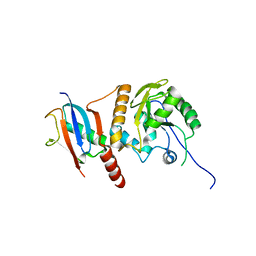 | | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase | | Descriptor: | G/T mismatch-specific thymine DNA glycosylase, Ubiquitin-like protein SMT3B | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-07-26 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase
J.Mol.Biol., 359, 2006
|
|
1X0S
 
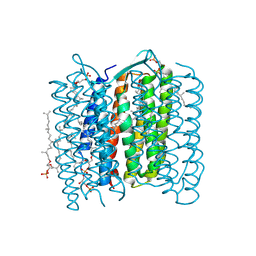 | | Crystal structure of the 13-cis isomer of bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nishikawa, T, Murakami, M, Kouyama, T. | | Deposit date: | 2005-03-28 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the 13-cis isomer of bacteriorhodopsin in the dark-adapted state.
J.Mol.Biol., 352, 2005
|
|
1WR1
 
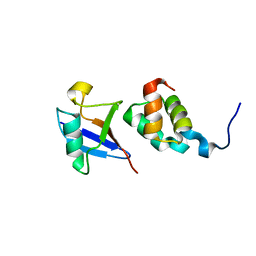 | | The complex structure of Dsk2p UBA with ubiquitin | | Descriptor: | Ubiquitin, Ubiquitin-like protein DSK2 | | Authors: | Ohno, A, Jee, J.G, Fujiwara, K, Tenno, T, Goda, N, Tochio, H, Hiroaki, H, kobayashi, H, Shirakawa, M. | | Deposit date: | 2004-10-08 | | Release date: | 2005-04-19 | | Last modified: | 2023-09-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition.
Structure, 13, 2005
|
|
1WR0
 
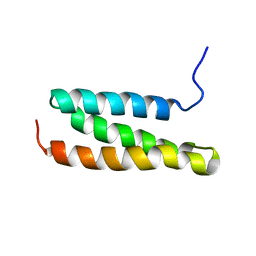 | | Structural characterization of the MIT domain from human Vps4b | | Descriptor: | SKD1 protein | | Authors: | Takasu, H, Jee, J.G, Ohno, A, Goda, N, Fujiwara, K, Tochio, H, Shirakawa, M, Hiroaki, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-07 | | Release date: | 2005-08-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the MIT domain from human Vps4b
Biochem.Biophys.Res.Commun., 334, 2005
|
|
2D2P
 
 | | The solution structure of micelle-bound peptide | | Descriptor: | Pituitary adenylate cyclase activating polypeptide-38 | | Authors: | Tateishi, Y, Jee, J.G, Inooka, H, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-09-14 | | Release date: | 2006-09-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The solution structure of micelle-bound peptide
To be Published
|
|
