7A20
 
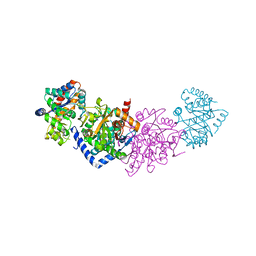 | | Azobenzene-Based Inhibitors for Tryptophan Synthase | | Descriptor: | SODIUM ION, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, Tryptophan synthase alpha chain,Tryptophan synthase beta chain | | Authors: | Rajendran, C, Sterner, R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Towards Photochromic Azobenzene-Based Inhibitors for Tryptophan Synthase.
Chemistry, 27, 2021
|
|
8S8S
 
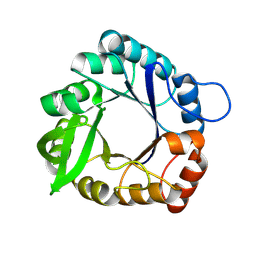 | | An induced-fit motion of a mobile loop | | Descriptor: | Imidazole glycerol phosphate synthase subunit HisF | | Authors: | Rajendran, C. | | Deposit date: | 2024-03-07 | | Release date: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.314 Å) | | Cite: | Conformational Modulation of a Mobile Loop Controls Catalysis in the ( beta alpha ) 8 -Barrel Enzyme of Histidine Biosynthesis HisF.
Jacs Au, 4, 2024
|
|
3O5T
 
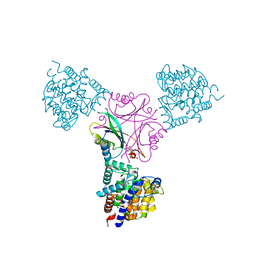 | | Structure of DraG-GlnZ complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dinitrogenase reductase activacting glicohydrolase, MAGNESIUM ION, ... | | Authors: | Rajendran, C, Li, X.-D, Winkler, F.K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of the GlnZ-DraG complex reveals a different form of PII-target interaction
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8S8R
 
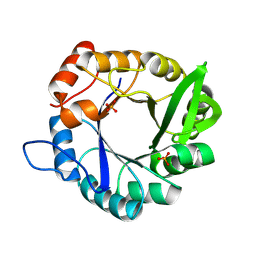 | | An induced-fit motion of a mobile loop | | Descriptor: | Imidazole glycerol phosphate synthase subunit HisF, PHOSPHATE ION | | Authors: | Rajendran, C. | | Deposit date: | 2024-03-07 | | Release date: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.196 Å) | | Cite: | Conformational Modulation of a Mobile Loop Controls Catalysis in the ( beta alpha ) 8 -Barrel Enzyme of Histidine Biosynthesis HisF.
Jacs Au, 4, 2024
|
|
8BLN
 
 | | Structure of RutB | | Descriptor: | Ureidoacrylate amidohydrolase RutB | | Authors: | Rajendran, C. | | Deposit date: | 2022-11-09 | | Release date: | 2023-09-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and Functional Characterization of the Ureidoacrylate Amidohydrolase RutB from Escherichia coli .
Biochemistry, 62, 2023
|
|
8BKD
 
 | | structure of RutB | | Descriptor: | N-(AMINOCARBONYL)-BETA-ALANINE, Ureidoacrylate amidohydrolase RutB | | Authors: | Rajendran, C. | | Deposit date: | 2022-11-09 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Characterization of the Ureidoacrylate Amidohydrolase RutB from Escherichia coli .
Biochemistry, 62, 2023
|
|
8BLM
 
 | | Structure of RutB | | Descriptor: | Ureidoacrylate amidohydrolase RutB | | Authors: | Rajendran, C. | | Deposit date: | 2022-11-09 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Ureidoacrylate Amidohydrolase RutB from Escherichia coli .
Biochemistry, 62, 2023
|
|
8BLL
 
 | | Structure of RutB | | Descriptor: | ACETATE ION, Ureidoacrylate amidohydrolase RutB | | Authors: | Rajendran, C. | | Deposit date: | 2022-11-09 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural and Functional Characterization of the Ureidoacrylate Amidohydrolase RutB from Escherichia coli .
Biochemistry, 62, 2023
|
|
3M97
 
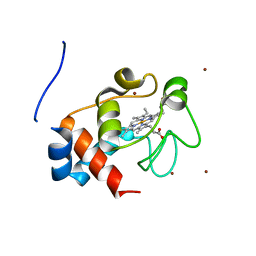 | | Structure of the soluble domain of cytochrome c552 with its flexible linker segment from Paracoccus denitrificans | | Descriptor: | Cytochrome c-552, HEME C, ZINC ION | | Authors: | Rajendran, C, Ermler, U, Ludwig, B, Michel, H. | | Deposit date: | 2010-03-20 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Structure at 1.5 A resolution of cytochrome c(552) with its flexible linker segment, a membrane-anchored protein from Paracoccus denitrificans.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4MM1
 
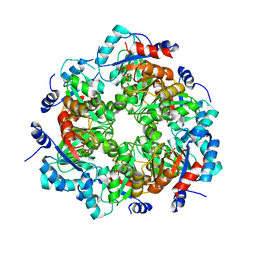 | | GGGPS from Methanothermobacter thermautotrophicus | | Descriptor: | Geranylgeranylglyceryl phosphate synthase, SN-GLYCEROL-1-PHOSPHATE, TRIETHYLENE GLYCOL | | Authors: | Rajendran, C, Peterhoff, D, Beer, B, Kumpula, E.P, Kapetaniou, E, Guldan, H, Wierenga, R.K, Sterner, R, Babinger, P. | | Deposit date: | 2013-09-07 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8004 Å) | | Cite: | A comprehensive analysis of the geranylgeranylglyceryl phosphate synthase enzyme family identifies novel members and reveals mechanisms of substrate specificity and quaternary structure organization.
Mol.Microbiol., 92, 2014
|
|
8BYW
 
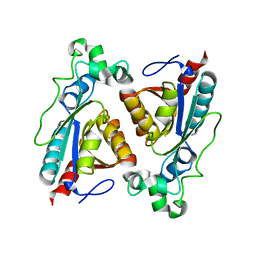 | | Rut B structure | | Descriptor: | Ureidoacrylate amidohydrolase RutB | | Authors: | Rajendran, C, Sterner, R, Busch, M. | | Deposit date: | 2022-12-14 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural and Functional Characterization of the Ureidoacrylate Amidohydrolase RutB from Escherichia coli .
Biochemistry, 62, 2023
|
|
4QO5
 
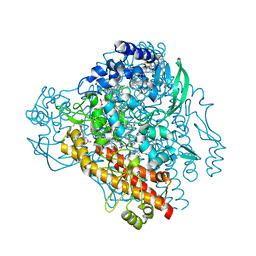 | | Hypothetical multiheme protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HEME C, ... | | Authors: | Rajendran, C. | | Deposit date: | 2014-06-19 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | In meso crystal structure of a novel membrane-associated octaheme cytochrome c from the Crenarchaeon Ignicoccus hospitalis.
Febs J., 283, 2016
|
|
4J2M
 
 | |
4FX7
 
 | |
6QUR
 
 | |
5NDY
 
 | |
6YMU
 
 | | Imidazole Glycerol Phosphate Synthase | | Descriptor: | Imidazole glycerol phosphate synthase subunit HisF, Imidazole glycerol phosphate synthase subunit HisH | | Authors: | Sterner, R, Rajendran, C, Andrea, K. | | Deposit date: | 2020-04-09 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Significance of the Protein Interface Configuration for Allostery in Imidazole Glycerol Phosphate Synthase.
Biochemistry, 59, 2020
|
|
5IR6
 
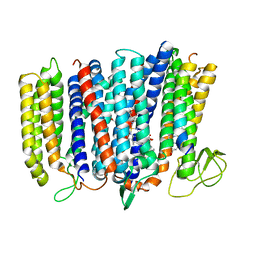 | | The structure of bd oxidase from Geobacillus thermodenitrificans | | Descriptor: | Bd-type quinol oxidase subunit I, Bd-type quinol oxidase subunit II, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, ... | | Authors: | Safarian, S, Mueller, H, Rajendran, C, Preu, J, Ovchinnikov, S, Kusumoto, T, Hirose, T, Langer, J, Sakamoto, J, Michel, H. | | Deposit date: | 2016-03-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases.
Science, 352, 2016
|
|
1W2L
 
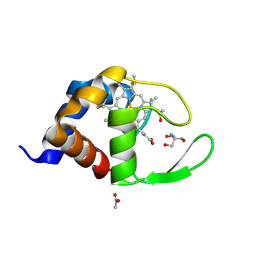 | | Cytochrome c domain of caa3 oxygen oxidoreductase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CYTOCHROME OXIDASE SUBUNIT II, ... | | Authors: | Srinivasan, V, Rajendran, C, Sousa, F.L, Melo, A.M.P, Saraiva, L.M, Pereira, M.M, Santana, M, Teixeira, M, Michel, H. | | Deposit date: | 2004-07-06 | | Release date: | 2005-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure at 1.3 A resolution of Rhodothermus marinus caa(3) cytochrome c domain.
J. Mol. Biol., 345, 2005
|
|
6RTZ
 
 | |
5NF1
 
 | |
5NEZ
 
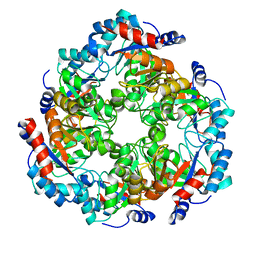 | |
6RU0
 
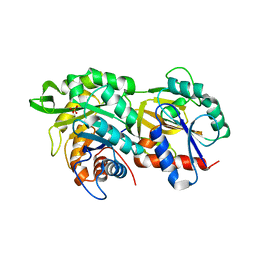 | |
5DOQ
 
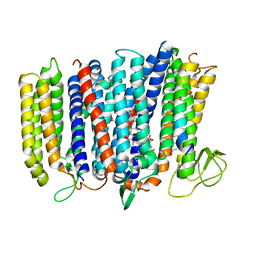 | | The structure of bd oxidase from Geobacillus thermodenitrificans | | Descriptor: | Bd-type quinol oxidase subunit I, Bd-type quinol oxidase subunit II, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, ... | | Authors: | Safarian, S, Mueller, H, Rajendran, C, Preu, J, Ovchinnikov, S, Kusumoto, T, Hirose, T, Langer, J, Sakamoto, J, Michel, H. | | Deposit date: | 2015-09-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases.
Science, 352, 2016
|
|
4NAF
 
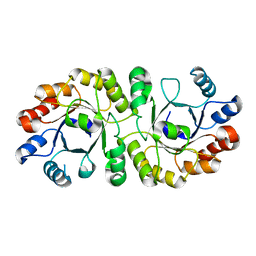 | | PrcB from Geobacillus kaustophilus, apo structure | | Descriptor: | Heptaprenylglyceryl phosphate synthase | | Authors: | Peterhoff, D, Beer, B, Rajendran, C, Kumpula, E.P, Kapetaniou, E, Guldan, H, Wierenga, R.K, Sterner, R, Babinger, P. | | Deposit date: | 2013-10-22 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A comprehensive analysis of the geranylgeranylglyceryl phosphate synthase enzyme family identifies novel members and reveals mechanisms of substrate specificity and quaternary structure organization.
Mol.Microbiol., 92, 2014
|
|
