5HM5
 
 | |
4GFJ
 
 | | Crystal structure of Topo-78, an N-terminal 78kDa fragment of topoisomerase V | | Descriptor: | GLYCEROL, Topoisomerase V, ZINC ION | | Authors: | Rajan, R, Prasad, R, Taneja, B, Wilson, S.H, Mondragon, A. | | Deposit date: | 2012-08-03 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Identification of one of the apurinic/apyrimidinic lyase active sites of topoisomerase V by structural and functional studies.
Nucleic Acids Res., 41, 2013
|
|
3M7G
 
 | |
3M6K
 
 | |
3M6Z
 
 | | Crystal structure of an N-terminal 44 kDa fragment of topoisomerase V in the presence of guanidium hydrochloride | | Descriptor: | CHLORIDE ION, GUANIDINE, MAGNESIUM ION, ... | | Authors: | Rajan, R, Taneja, B, Mondragon, A. | | Deposit date: | 2010-03-16 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of minimal catalytic fragments of topoisomerase v reveals conformational changes relevant for DNA binding.
Structure, 18, 2010
|
|
3M7D
 
 | |
1YCL
 
 | | Crystal Structure of B. subtilis LuxS in Complex with a Catalytic 2-Ketone Intermediate | | Descriptor: | (S)-2-AMINO-4-[(2S,3R)-2,3,5-TRIHYDROXY-4-OXO-PENTYL]MERCAPTO-BUTYRIC ACID, COBALT (II) ION, S-ribosylhomocysteinase, ... | | Authors: | Rajan, R, Zhu, J, Hu, X, Pei, D, Bell, C.E. | | Deposit date: | 2004-12-22 | | Release date: | 2005-03-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of S-Ribosylhomocysteinase (LuxS) in Complex with a Catalytic 2-Ketone Intermediate.
Biochemistry, 44, 2005
|
|
1RBE
 
 | |
1RBF
 
 | |
1RBI
 
 | |
1RBC
 
 | |
1RBG
 
 | |
1RBD
 
 | |
1RBH
 
 | |
1XP8
 
 | | Deinococcus radiodurans RecA in complex with ATP-gamma-S | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, RecA protein | | Authors: | Bell, C.E, Rajan, R. | | Deposit date: | 2004-10-08 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of RecA from Deinococcus radiodurans: insights into the structural basis of extreme radioresistance.
J.Mol.Biol., 344, 2004
|
|
2FQT
 
 | | Crystal structure of B.subtilis LuxS in complex with (2S)-2-Amino-4-[(2R,3S)-2,3-dihydroxy-3-N-hydroxycarbamoyl-propylmercapto]butyric acid | | Descriptor: | (2S)-2-AMINO-4-[(2R,3S)-2,3-DIHYDROXY-3-N-HYDROXYCARBAMOYL-PROPYLMERCAPTO]BUTYRIC ACID, COBALT (II) ION, S-ribosylhomocysteine lyase, ... | | Authors: | Shen, G, Rajan, R, Zhu, J, Bell, C.E, Pei, D. | | Deposit date: | 2006-01-18 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Design and Synthesis of Substrate Analogue Inhibitors of S-Ribosylhomocysteinase (LuxS)
J.Med.Chem., 49, 2006
|
|
2FQO
 
 | | Crystal structure of B. subtilis LuxS in complex with (2S)-2-Amino-4-[(2R,3R)-2,3-dihydroxy-3-N- hydroxycarbamoyl-propylmercapto]butyric acid | | Descriptor: | (2S)-2-AMINO-4-[(2R,3R)-2,3-DIHYDROXY-3-N-HYDROXYCARBAMOYL-PROPYLMERCAPTO]BUTYRIC ACID, COBALT (II) ION, S-ribosylhomocysteine lyase, ... | | Authors: | Shen, G, Rajan, R, Zhu, J, Bell, C.E, Pei, D. | | Deposit date: | 2006-01-18 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Design and Synthesis of Substrate and Intermediate Analogue Inhibitors of S-Ribosylhomocysteinase
J.Med.Chem., 49, 2006
|
|
7X7N
 
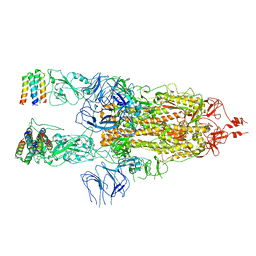 | | 3D model of the 3-RBD up single trimeric spike protein of SARS-CoV2 in the presence of synthetic peptide SIH-5. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, Synthetic peptide SIH-5 | | Authors: | Khatri, B, Pramanick, I, Malladi, S.K, Rajmani, R.S, Kumar, S, Ghosh, P, Sengupta, N, Rahisuddin, R, Kumaran, S, Ringe, R.P, Varadarajan, R, Dutta, S, Chatterjee, J. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | A dimeric proteomimetic prevents SARS-CoV-2 infection by dimerizing the spike protein.
Nat.Chem.Biol., 18, 2022
|
|
1A19
 
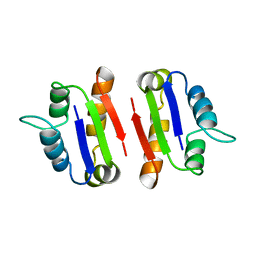 | | BARSTAR (FREE), C82A MUTANT | | Descriptor: | BARSTAR | | Authors: | Ratnaparkhi, G.S, Varadarajan, R. | | Deposit date: | 1997-12-25 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discrepancies between the NMR and X-ray structures of uncomplexed barstar: analysis suggests that packing densities of protein structures determined by NMR are unreliable.
Biochemistry, 37, 1998
|
|
7EPI
 
 | |
7EPG
 
 | |
7EPJ
 
 | |
5FCE
 
 | |
6IEP
 
 | | GAPDH of Streptococcus agalactiae | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Ponnuraj, K, Nagarajan, R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of GAPDH of Streptococcus agalactiae and characterization of its interaction with extracellular matrix molecules
Microb. Pathog., 127, 2018
|
|
1D5D
 
 | |
