4YGI
 
 | |
2QXU
 
 | |
1T4M
 
 | |
2QXT
 
 | |
1T2N
 
 | |
3SOW
 
 | |
3SOU
 
 | |
3SOX
 
 | | Structure of UHRF1 PHD finger in the free form | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Rajakumara, E, Patel, D.J. | | Deposit date: | 2011-06-30 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6501 Å) | | Cite: | PHD Finger Recognition of Unmodified Histone H3R2 Links UHRF1 to Regulation of Euchromatic Gene Expression.
Mol.Cell, 43, 2011
|
|
8WKL
 
 | |
8H8P
 
 | | Crystal structure of thiomorpholine-carboxylate dehydrogenase from Candida parapsilosis. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Uma Mahesh, M.V.N, Abhishek, S, Faidh, M.A, Rajakumara, E, Chadha, A. | | Deposit date: | 2022-10-24 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of a Ornithine cyclodeaminase/Mu-crystallin homolog purified from Candida parapsilosis ATCC 7330.
To Be Published
|
|
7DUF
 
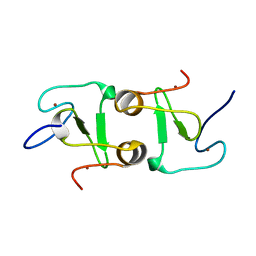 | | Crystal structure of VIM1 PHD finger. | | Descriptor: | E3 ubiquitin-protein ligase ORTHRUS 2, ZINC ION | | Authors: | Abhishek, S, Deeksha, W, Patel, D.J, Rajakumara, E. | | Deposit date: | 2021-01-08 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Helical and beta-Turn Conformations in the Peptide Recognition Regions of the VIM1 PHD Finger Abrogate H3K4 Peptide Recognition.
Biochemistry, 60, 2021
|
|
6M2V
 
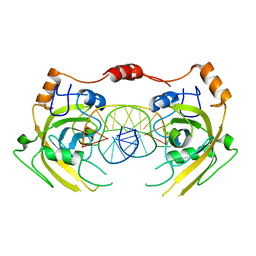 | | Crystal structure of UHRF1 SRA complexed with fully-mCHG DNA. | | Descriptor: | DNA (5'-D(*TP*CP*AP*CP*GP*(5CM)P*TP*GP*CP*GP*TP*GP*A)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Abhishek, S, Nakarakanti, N.K, Deeksha, W, Rajakumara, E. | | Deposit date: | 2020-03-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanistic insights into recognition of symmetric methylated cytosines in CpG and non-CpG DNA by UHRF1 SRA.
Int.J.Biol.Macromol., 170, 2021
|
|
3E53
 
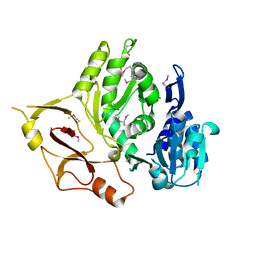 | |
1U2P
 
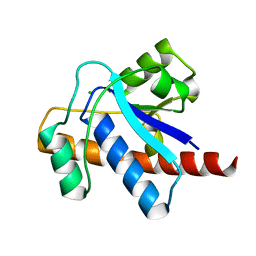 | | Crystal structure of Mycobacterium tuberculosis Low Molecular Protein Tyrosine Phosphatase (MPtpA) at 1.9A resolution | | Descriptor: | CHLORIDE ION, low molecular weight protein-tyrosine-phosphatase | | Authors: | Madhurantakam, C, Rajakumara, E, Mazumdar, P.A, Saha, B, Mitra, D, Wiker, H.G, Sankaranarayanan, R, Das, A.K. | | Deposit date: | 2004-07-20 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Low-Molecular-Weight Protein Tyrosine Phosphatase from Mycobacterium tuberculosis at 1.9-A Resolution
J.Bacteriol., 187, 2005
|
|
1U2Q
 
 | | Crystal structure of Mycobacterium tuberculosis Low Molecular Weight Protein Tyrosine Phosphatase (MPtpA) at 2.5A resolution with glycerol in the active site | | Descriptor: | CHLORIDE ION, GLYCEROL, low molecular weight protein-tyrosine-phosphatase | | Authors: | Madhurantakam, C, Rajakumara, E, Mazumdar, P.A, Saha, B, Mitra, D, Wiker, H.G, Sankaranarayanan, R, Das, A.K. | | Deposit date: | 2004-07-20 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Low-Molecular-Weight Protein Tyrosine Phosphatase from Mycobacterium tuberculosis at 1.9-A Resolution
J.Bacteriol., 187, 2005
|
|
