2G44
 
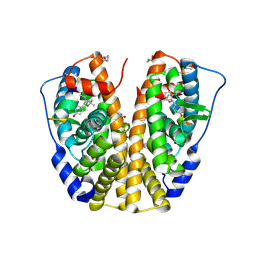 | | Human Estrogen Receptor Alpha Ligand-Binding Domain In Complex With OBCP-1M-G and A Glucocorticoid Receptor Interacting Protein 1 NR Box II Peptide | | Descriptor: | 4-[(1S,2R,5S)-4,4,8-TRIMETHYL-3-OXABICYCLO[3.3.1]NON-7-EN-2-YL]PHENOL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Greene, G.L. | | Deposit date: | 2006-02-21 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and characterization of novel estrogen
receptor agonist ligands and development of biochips for
nuclear receptor drug discovery
Thesis, 2006
|
|
4NQE
 
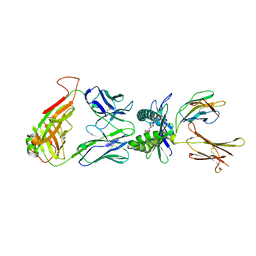 | | Crystal structure of TCR-MR1 ternary complex bound to 5-(2-oxoethylideneamino)-6-D-ribitylaminouracil | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-(2-oxoethylidene)amino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-04-16 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | T-cell activation by transitory neo-antigens derived from distinct microbial pathways.
Nature, 509, 2014
|
|
1DZ1
 
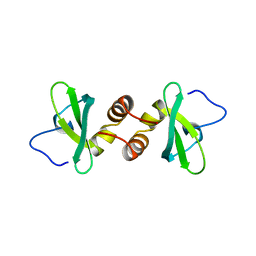 | | Mouse HP1 (M31) C terminal (shadow chromo) domain | | Descriptor: | MODIFIER 1 PROTEIN | | Authors: | Brasher, S.V, Smith, B.O, Fogh, R.H, Nietlispach, D, Thiru, A, Nielsen, P.R, Broadhurst, R.W, Ball, L.J, Murzina, N, Laue, E.D. | | Deposit date: | 2000-02-11 | | Release date: | 2000-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of Mouse Hp1 Suggests a Unique Mode of Single Peptide Recognition by the Shadow Chromo Domain Dimer
Embo J., 19, 2000
|
|
1E04
 
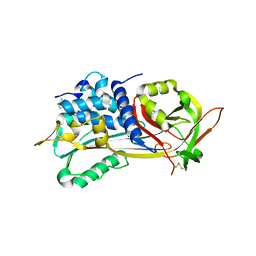 | | PLASMA BETA ANTITHROMBIN-III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, GLYCEROL, ... | | Authors: | Mccoy, A.J, Skinner, R, Abrahams, J.-P, Pei, X.Y, Carrell, R.W. | | Deposit date: | 2000-03-09 | | Release date: | 2000-06-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Beta-Antithrombin and the Effect of Glycosylation on Antithrombin'S Heparin Affinity and Activity.
J.Mol.Biol., 326, 2003
|
|
1GYG
 
 | |
1GS6
 
 | | Crystal structure of M144A mutant of Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, MAGNESIUM ION | | Authors: | Ellis, M.J, Prudencio, M, Dodd, F.E, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Crystallographic Studies of the met144Ala, Asp92Asn and His254Phe Mutants of the Nitrite Reductase from Alcaligenes Xylosoxidans Provide Insight Into the Enzyme Mechanism.
J.Mol.Biol., 316, 2002
|
|
1FEJ
 
 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-21 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
6X63
 
 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UX8
 
 | |
1FG6
 
 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-25 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
1YC6
 
 | |
1FF0
 
 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES. | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-24 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
5TB7
 
 | | Structure of nitrite reductase AniA from Neisseria gonorrhoeae, space group P212121 | | Descriptor: | COPPER (II) ION, Nitrite reductase, PHOSPHATE ION | | Authors: | Hamza, A, Williamson, Z.A, Reed, R.W, Sikora, A.E, Korotkov, K.V. | | Deposit date: | 2016-09-11 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Peptide Inhibitors Targeting the Neisseria gonorrhoeae Pivotal Anaerobic Respiration Factor AniA.
Antimicrob. Agents Chemother., 61, 2017
|
|
4NQD
 
 | | Crystal structure of TCR-MR1 ternary complex and non-covalently bound 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-(2-oxopropylidene)amino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-04-16 | | Last modified: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | T-cell activation by transitory neo-antigens derived from distinct microbial pathways.
Nature, 509, 2014
|
|
4L4T
 
 | | Structure of human MAIT TCR in complex with human MR1-6-FP | | Descriptor: | 2-amino-4-oxo-3,4-dihydropteridine-6-carbaldehyde, Beta-2-microglobulin, MAIT T-cell receptor alpha chain, ... | | Authors: | Patel, O, Kjer-Nielsen, L, Le Nours, J, Eckle, S.B.G, Birkinshaw, R.W, Beddoe, T, Corbett, A.J, Liu, L, Miles, J.J, Meehan, B, Reantragoon, R, Sandoval-Romero, M.L, Sullivan, L.C, Brooks, A.G, Chen, Z, Fairlie, D.P, McCluskey, J, Rossjohn, J. | | Deposit date: | 2013-06-09 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of vitamin B metabolites by mucosal-associated invariant T cells.
Nat Commun, 4, 2013
|
|
6V9H
 
 | | Ankyrin repeat and SOCS-box protein 9 (ASB9), ElonginB (ELOB), and ElonginC (ELOC) bound to its substrate Brain-type Creatine Kinase (CKB) | | Descriptor: | Ankyrin repeat and SOCS box protein 9, Creatine kinase B-type, Elongin-B, ... | | Authors: | Komives, E.A, Lumpkin, R.J, Baker, R.W, Leschziner, A.E. | | Deposit date: | 2019-12-13 | | Release date: | 2020-04-29 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and dynamics of the ASB9 CUL-RING E3 Ligase.
Nat Commun, 11, 2020
|
|
5VHU
 
 | | E. coli CFT073 c3406 | | Descriptor: | Isomerase, SULFATE ION | | Authors: | Cech, D.L, Pratt, A.C, Woodard, R.W. | | Deposit date: | 2017-04-13 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of arabinose-5-phosphate isomerases.
To Be Published
|
|
1G8E
 
 | | CRYSTAL STRUCTURE OF FLHD FROM ESCHERICHIA COLI | | Descriptor: | FLAGELLAR TRANSCRIPTIONAL ACTIVATOR FLHD | | Authors: | Campos, A, Zhang, R.G, Alkire, R.W, Matsumura, P, Westbrook, E.M. | | Deposit date: | 2000-11-17 | | Release date: | 2001-03-07 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the global regulator FlhD from Escherichia coli at 1.8 A resolution.
Mol.Microbiol., 39, 2001
|
|
4L4V
 
 | | Structure of human MAIT TCR in complex with human MR1-RL-6-Me-7-OH | | Descriptor: | 1-deoxy-1-(7-hydroxy-6-methyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Patel, O, Kjer-Nielsen, L, Le Nours, J, Eckle, S.B.G, Birkinshaw, R.W, Beddoe, T, Corbett, A.J, Liu, L, Miles, J.J, Meehan, B, Reantragoon, R, Sandoval-Romero, M.L, Sullivan, L.C, Brooks, A.G, Chen, Z, Fairlie, D.P, McCluskey, J, Rossjohn, J. | | Deposit date: | 2013-06-09 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition of vitamin B metabolites by mucosal-associated invariant T cells.
Nat Commun, 4, 2013
|
|
6V9I
 
 | | cryo-EM structure of Cullin5 bound to RING-box protein 2 (Cul5-Rbx2) | | Descriptor: | Immunoglobulin G-binding protein G,Cullin-5, RING-box protein 2, ZINC ION | | Authors: | Komives, E.A, Lumpkin, R.J, Baker, R.W, Leschziner, A.E. | | Deposit date: | 2019-12-13 | | Release date: | 2020-04-29 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structure and dynamics of the ASB9 CUL-RING E3 Ligase.
Nat Commun, 11, 2020
|
|
1GS8
 
 | | Crystal structure of mutant D92N Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Ellis, M.J, Prudencio, M, Dodd, F.E, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Crystallographic Studies of the met144Ala, Asp92Asn and His254Phe Mutants of the Nitrite Reductase from Alcaligenes Xylosoxidans Provide Insight Into the Enzyme Mechanism.
J.Mol.Biol., 316, 2002
|
|
5UQI
 
 | |
1FFI
 
 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-25 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
1FGC
 
 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-28 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
6XSU
 
 | | GH5-4 broad specificity endoglucanase from Ruminococcus flavefaciens | | Descriptor: | ETHANOL, GH5-4 broad specificity endoglucanase | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-07-16 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
