3J9F
 
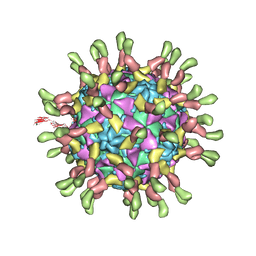 | | Poliovirus complexed with soluble, deglycosylated poliovirus receptor (Pvr) at 4 degrees C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-02-11 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
3K0X
 
 | | Crystal structure of telomere capping protein Ten1 from Saccharomyces pombe | | Descriptor: | IODIDE ION, Protein Ten1 | | Authors: | Gelinas, A.D, Reyes, F.E, Batey, R.T, Wuttke, D.S. | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3MMP
 
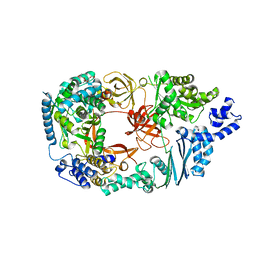 | | Structure of the Qb replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Elongation factor Tu 2, Elongation factor Ts, ... | | Authors: | Kidmose, R.T, Vasiliev, N.N, Chetverin, A.B, Knudsen, C.R, Andersen, G.R. | | Deposit date: | 2010-04-20 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1TNS
 
 | |
1TNT
 
 | |
4R0D
 
 | | Crystal structure of a eukaryotic group II intron lariat | | Descriptor: | GROUP IIB INTRON LARIAT, IRIDIUM HEXAMMINE ION, LIGATED EXONS, ... | | Authors: | Robart, A.R, Chan, R.T, Peters, J.K, Rajashankar, K.R, Toor, N. | | Deposit date: | 2014-07-30 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.676 Å) | | Cite: | Crystal structure of a eukaryotic group II intron lariat.
Nature, 514, 2014
|
|
6P2H
 
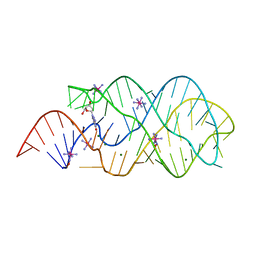 | |
6PP7
 
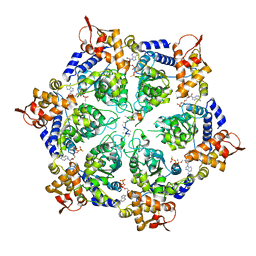 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6POD
 
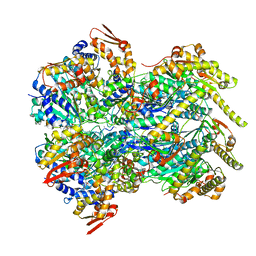 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PP5
 
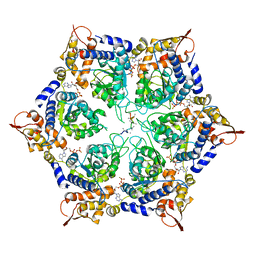 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PO3
 
 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PPE
 
 | | ClpP and ClpX IGF loop in ClpX-ClpP complex with D7 symmetry | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6POS
 
 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PP6
 
 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PO1
 
 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PP8
 
 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
1X6X
 
 | |
1X6Y
 
 | |
6PXI
 
 | | The crystal structure of a singly capped HslUV complex with an axial pore plug and a HslU E257Q mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV, ... | | Authors: | Baytshtok, V, Grant, R.A, Sauer, R.T. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.447 Å) | | Cite: | Heat activates the AAA+ HslUV protease by melting an axial autoinhibitory plug.
Cell Rep, 34, 2021
|
|
1X6P
 
 | |
6PVU
 
 | | RNase A in complex with hexametaphosphate | | Descriptor: | 2,4,6,8,10,12-hexahydroxy-2lambda~5~,4lambda~5~,6lambda~5~,8lambda~5~,10lambda~5~,12lambda~5~-cyclohexaphosphoxane-2,4,6,8,10,12-hexone, Ribonuclease pancreatic | | Authors: | Windsor, I.W, Sheppard, S.M, Cummins, C.C, Raines, R.T. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Nucleoside Tetra- and Pentaphosphates Prepared Using a Tetraphosphorylation Reagent Are Potent Inhibitors of Ribonuclease A.
J.Am.Chem.Soc., 141, 2019
|
|
6PVW
 
 | | RNase A in complex with cp4pA | | Descriptor: | 2,4,6,8-tetrahydroxy-1,3,5,7,2lambda~5~,4lambda~5~,6lambda~5~,8lambda~5~-tetroxatetraphosphocane-2,4,6,8-tetrone, 5'-O-[(R)-hydroxy{[(4R,8S)-4,6,8-trihydroxy-2,4,6,8-tetraoxo-1,3,5,7,2lambda~5~,4lambda~5~,6lambda~5~,8lambda~5~-tetroxatetraphosphocan-2-yl]oxy}phosphoryl]adenosine, Ribonuclease pancreatic | | Authors: | Windsor, I.W, Sheppard, S.M, Cummins, C.C, Raines, R.T. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleoside Tetra- and Pentaphosphates Prepared Using a Tetraphosphorylation Reagent Are Potent Inhibitors of Ribonuclease A.
J.Am.Chem.Soc., 141, 2019
|
|
1VG3
 
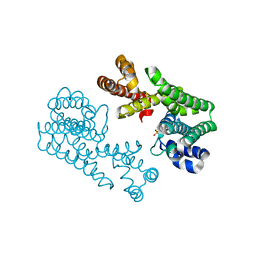 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima A76Y/S77F mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
1VG7
 
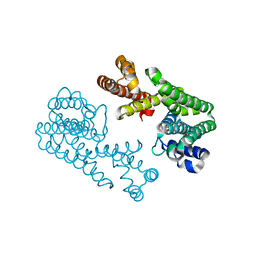 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima F132A/L128A/I123A/D62A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
1X6Z
 
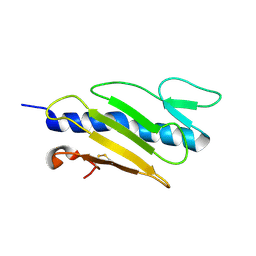 | |
