7VQ6
 
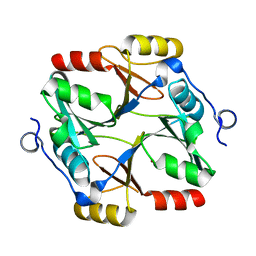 | | Structure of a specialized glyoxalase from Gossypium hirsutum | | Descriptor: | Lactoylglutathione lyase, NICKEL (II) ION | | Authors: | Li, H, Hu, Y.M, Dai, L.H, Chen, C.C, Huang, J.W, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure and biochemical analysis of the specialized deoxynivalenol-detoxifying glyoxalase SPG from Gossypium hirsutum.
Int.J.Biol.Macromol., 200, 2022
|
|
7UYU
 
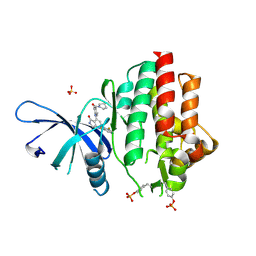 | | Crystal structure of TYK2 kinase domain in complex with compound 30 | | Descriptor: | 2-(2,6-difluorophenyl)-4-[4-(pyrrolidine-1-carbonyl)anilino]-5H-pyrrolo[3,4-b]pyridin-5-one, Non-receptor tyrosine-protein kinase TYK2, SULFATE ION | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYR
 
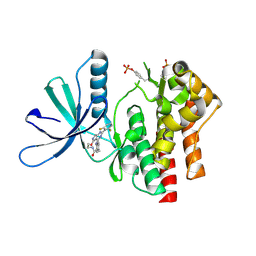 | | Crystal structure of TYK2 kinase domain in complex with compound 12 | | Descriptor: | 2-(4-{[2-(2,6-difluorophenyl)-5-oxo-5H-pyrrolo[3,4-d]pyrimidin-4-yl]amino}phenyl)-N-ethylacetamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYT
 
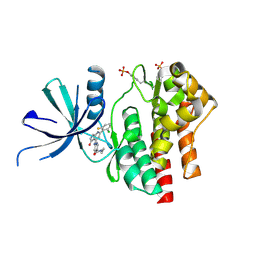 | | Crystal structure of TYK2 kinase domain in complex with compound 25 | | Descriptor: | 6-{[(2M)-2-(2-chloro-6-fluorophenyl)-5-oxo-5H-pyrrolo[3,4-b]pyridin-4-yl]amino}-N-ethylpyridine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYW
 
 | | Crystal structure of JAK2 kinase domain in complex with compound 30 | | Descriptor: | 2-(2,6-difluorophenyl)-4-[4-(pyrrolidine-1-carbonyl)anilino]-5H-pyrrolo[3,4-b]pyridin-5-one, Tyrosine-protein kinase JAK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYV
 
 | | Crystal structure of JAK3 kinase domain in complex with compound 25 | | Descriptor: | 6-{[(2M)-2-(2-chloro-6-fluorophenyl)-5-oxo-5H-pyrrolo[3,4-b]pyridin-4-yl]amino}-N-ethylpyridine-3-carboxamide, CHLORIDE ION, Tyrosine-protein kinase JAK3 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYS
 
 | | Crystal structure of TYK2 kinase domain in complex with compound 16 | | Descriptor: | 2-(2,6-difluorophenyl)-4-(4-methoxyanilino)-5H-pyrrolo[3,4-d]pyrimidin-5-one, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UIY
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIW
 
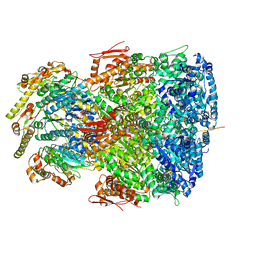 | | ClpAP complex bound to ClpS N-terminal extension, class IIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UJ0
 
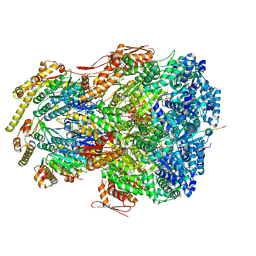 | | ClpAP complex bound to ClpS N-terminal extension, class IIIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIZ
 
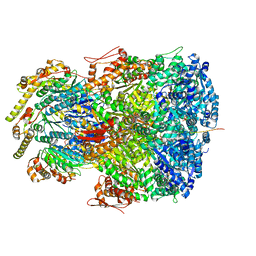 | | ClpAP complex bound to ClpS N-terminal extension, class IIc | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIX
 
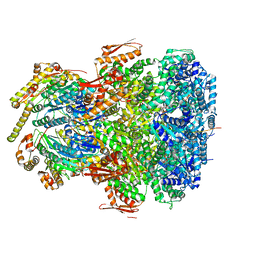 | | ClpAP complex bound to ClpS N-terminal extension, class I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W5S
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | Authors: | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7W5T
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7W5V
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
1BAZ
 
 | |
1BDV
 
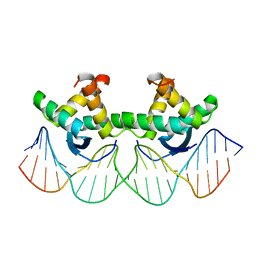 | | ARC FV10 COCRYSTAL | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*AP*TP*AP*GP*AP*AP*GP*CP*AP*CP*TP*CP*TP*AP*CP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*GP*TP*AP*GP*AP*GP*TP*GP*CP*TP*TP*CP*TP*AP*TP*CP*AP*T)-3'), PROTEIN (ARC FV10 REPRESSOR) | | Authors: | Schildbach, J.F, Karzai, A.W, Raumann, B.E, Sauer, R.T. | | Deposit date: | 1998-05-11 | | Release date: | 1999-01-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Origins of DNA-binding specificity: role of protein contacts with the DNA backbone.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1C8W
 
 | | THR45GLY VARIANT OF RIBONUCLEASE A | | Descriptor: | ACETATE ION, CHLORIDE ION, PROTEIN (Ribonuclease A) | | Authors: | Kelemen, B.R, Sweeney, R.T, Schultz, L.W, Raines, R.T. | | Deposit date: | 1999-07-30 | | Release date: | 2002-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Excavating an active site: the nucleobase specificity of ribonuclease A.
Biochemistry, 39, 2000
|
|
1WL2
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima R90A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1WL1
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima H74A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1WL0
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima R44A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1WL3
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima R91A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1WKZ
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima K41A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1VWD
 
 | | STREPTAVIDIN-CYCLO-AC-[CHPQFC]-NH2, PH 3.0 | | Descriptor: | PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
1VWP
 
 | |
