4JUN
 
 | | Crystal structure of H5N1 influenza virus hemagglutinin, clade 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | DuBois, R.M, Zaraket, H, Reddivari, M, Coop, T, Heath, R.J, White, S.W, Russell, C.J. | | Deposit date: | 2013-03-25 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3398 Å) | | Cite: | To be published
To be Published
|
|
2R5Q
 
 | | Crystal Structure Analysis of HIV-1 Subtype C Protease Complexed with Nelfinavir | | Descriptor: | 2-[2-HYDROXY-3-(3-HYDROXY-2-METHYL-BENZOYLAMINO)-4-PHENYL SULFANYL-BUTYL]-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID TERT-BUTYLAMIDE, Protease | | Authors: | Coman, R.M, Robbins, A.H, McKenna, R, Dunn, B.M. | | Deposit date: | 2007-09-04 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Contribution of Naturally Occurring Polymorphisms in Altering the Biochemical and Structural Characteristics of HIV-1 Subtype C Protease
Biochemistry, 47, 2008
|
|
4K1C
 
 | | VCX1 Calcium/Proton Exchanger | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, IODIDE ION, ... | | Authors: | Waight, A.B, Pedersen, B.P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2013-04-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for alternating access of a eukaryotic calcium/proton exchanger.
Nature, 499, 2013
|
|
4K2D
 
 | | Crystal structure of Burkholderia Pseudomallei DsbA | | Descriptor: | GLYCEROL, Thiol:disulfide interchange protein | | Authors: | McMahon, R.M. | | Deposit date: | 2013-04-09 | | Release date: | 2013-08-14 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Disarming Burkholderia pseudomallei: Structural and Functional Characterization of a Disulfide Oxidoreductase (DsbA) Required for Virulence In Vivo.
Antioxid Redox Signal, 20, 2014
|
|
1S8J
 
 | | Nitrate-bound D85S mutant of bacteriorhodopsin | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin precursor, NITRATE ION, ... | | Authors: | Facciotti, M.T, Cheung, V.S, Lunde, C.S, Rouhani, S, Baliga, N.S, Glaeser, R.M. | | Deposit date: | 2004-02-02 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Specificity of anion binding in the substrate pocket of bacteriorhodopsin.
Biochemistry, 43, 2004
|
|
1IIR
 
 | | Crystal Structure of UDP-glucosyltransferase GtfB | | Descriptor: | MAGNESIUM ION, SULFATE ION, glycosyltransferase GtfB | | Authors: | Mulichak, A.M, Losey, H.C, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2001-04-24 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics.
Structure, 9, 2001
|
|
2R8N
 
 | | Structural Analysis of the Unbound Form of HIV-1 Subtype C Protease | | Descriptor: | GLYCEROL, Pol protein | | Authors: | Coman, R.M, Robbins, A.H, McKenna, R, Dunn, B.M. | | Deposit date: | 2007-09-11 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution structure of unbound human immunodeficiency virus 1 subtype C protease: implications of flap dynamics and drug resistance.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4K0E
 
 | | X-ray crystal structure of a heavy metal efflux pump, crystal form II | | Descriptor: | Heavy metal cation tricomponent efflux pump ZneA(CzcA-like), ZINC ION | | Authors: | Pak, J.E, Ngonlong Ekende, E, Vandenbussche, G, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2013-04-03 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.713 Å) | | Cite: | Structures of intermediate transport states of ZneA, a Zn(II)/proton antiporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K0J
 
 | | X-ray crystal structure of a heavy metal efflux pump, crystal form I | | Descriptor: | Heavy metal cation tricomponent efflux pump ZneA(CzcA-like), ZINC ION | | Authors: | Pak, J.E, Stroud, R.M, Ngonlong Ekende, E, Vandenbussche, G, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2013-04-04 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of intermediate transport states of ZneA, a Zn(II)/proton antiporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2L2N
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for the first dsRBD of protein HYL1 | | Descriptor: | Hyponastic leave 1 | | Authors: | Rasia, R.M, Mateos, J.L, Bologna, N.G, Burdisso, P, Imbert, L, Palatnik, J.F, Boisbouvier, J. | | Deposit date: | 2010-08-23 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and RNA Interactions of the Plant MicroRNA Processing-Associated Protein HYL1.
Biochemistry, 49, 2010
|
|
2LJC
 
 | |
2LCB
 
 | | Solution Structure of a Minor and Transiently Formed State of a T4 Lysozyme Mutant | | Descriptor: | Lysozyme | | Authors: | Bouvignies, G, Vallurupalli, P, Hansen, D, Correia, B, Lange, O, Bah, A, Vernon, R.M, Dahlquist, F.W, Baker, D, Kay, L.E. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a minor and transiently formed state of a T4 lysozyme mutant.
Nature, 477, 2011
|
|
1S8L
 
 | | Anion-free form of the D85S mutant of bacteriorhodopsin from crystals grown in the presence of halide | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin precursor, RETINAL | | Authors: | Facciotti, M.T, Cheung, V.S, Lunde, C.S, Rouhani, S, Baliga, N.S, Glaeser, R.M. | | Deposit date: | 2004-02-02 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Specificity of anion binding in the substrate pocket of bacteriorhodopsin.
Biochemistry, 43, 2004
|
|
2OYU
 
 | | Indomethacin-(S)-alpha-ethyl-ethanolamide bound to Cyclooxygenase-1 | | Descriptor: | 2-[1-(4-CHLOROBENZOYL)-5-METHOXY-2-METHYL-1H-INDOL-3-YL]-N-[(1S)-1-(HYDROXYMETHYL)PROPYL]ACETAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Harman, C.A, Garavito, R.M. | | Deposit date: | 2007-02-23 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of enantioselective inhibition of cyclooxygenase-1 by S-alpha-substituted indomethacin ethanolamides.
J.Biol.Chem., 282, 2007
|
|
4K5Y
 
 | | Crystal structure of human corticotropin-releasing factor receptor 1 (CRF1R) in complex with the antagonist CP-376395 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6-dimethyl-N-(pentan-3-yl)-2-(2,4,6-trimethylphenoxy)pyridin-4-amine, ... | | Authors: | Hollenstein, K, Kean, J, Bortolato, A, Cheng, R.K.Y, Dore, A.S, Jazayeri, A, Cooke, R.M, Weir, M, Marshall, F.H. | | Deposit date: | 2013-04-15 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.977 Å) | | Cite: | Structure of class B GPCR corticotropin-releasing factor receptor 1.
Nature, 499, 2013
|
|
2RKD
 
 | | The Structure of rat cytosolic PEPCK in complex with 3-phosphonopropionate | | Descriptor: | 3-PHOSPHONOPROPANOIC ACID, MANGANESE (II) ION, Phosphoenolpyruvate carboxykinase, ... | | Authors: | Sullivan, S.M, Stiffin, R.M, Carlson, G.M, Holyoak, T. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Inhibition of Cytosolic PEPCK by Substrate Analogues. Kinetic and Structural Characterization of Inhibitor Recognition.
Biochemistry, 47, 2008
|
|
2JQP
 
 | | NMR structure determination of Bungatoxin from Bungarus candidus (Malayan Krait) | | Descriptor: | Weak toxin 1 | | Authors: | Vivekanandan, S, Jois, S.D, Kini, R.M, Troncone, L.R.P, de Magalhaes, L, Ujikawa, G.Y, Ramos, A.T. | | Deposit date: | 2007-06-06 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution Structure of Bungatoxin from Bungarus Candidus (Malayan Krait)venom
To be Published
|
|
2RK8
 
 | | The Structure of rat cytosolic PEPCK in complex with phosphonoformate | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, MANGANESE (II) ION, ... | | Authors: | Sullivan, S.M, Stiffin, R.M, Carlson, G.M, Holyoak, T. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Inhibition of Cytosolic PEPCK by Substrate Analogues. Kinetic and Structural Characterization of Inhibitor Recognition.
Biochemistry, 47, 2008
|
|
2TGF
 
 | | THE SOLUTION STRUCTURE OF HUMAN TRANSFORMING GROWTH FACTOR ALPHA | | Descriptor: | TRANSFORMING GROWTH FACTOR-ALPHA | | Authors: | Harvey, T.S, Wilkinson, A.J, Tappin, M.J, Cooke, R.M, Campbell, I.D. | | Deposit date: | 1991-01-23 | | Release date: | 1993-04-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human transforming growth factor alpha.
Eur.J.Biochem., 198, 1991
|
|
2MFN
 
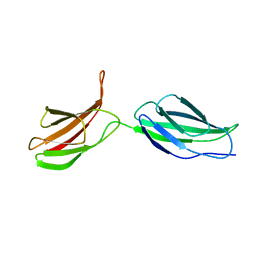 | | SOLUTION NMR STRUCTURE OF LINKED CELL ATTACHMENT MODULES OF MOUSE FIBRONECTIN CONTAINING THE RGD AND SYNERGY REGIONS, 10 STRUCTURES | | Descriptor: | FIBRONECTIN | | Authors: | Copie, V, Tomita, Y, Akiyama, S.K, Aota, S, Yamada, K.M, Venable, R.M, Pastor, R.W, Krueger, S, Torchia, D.A. | | Deposit date: | 1998-02-11 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of linked cell attachment modules of mouse fibronectin containing the RGD and synergy regions: comparison with the human fibronectin crystal structure.
J.Mol.Biol., 277, 1998
|
|
2BEW
 
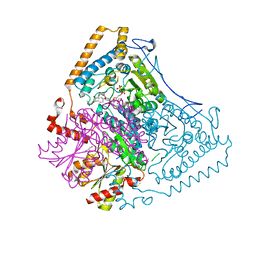 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXYPHENYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-30 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BFZ
 
 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH4.5 using 20mM ZnSO4 in buffer. 1mM DTT was used as a reducing agent. Cys221 is oxidized. | | Descriptor: | AZIDE ION, BETA-LACTAMASE II, GLYCEROL, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-16 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism
Biochemistry, 44, 2005
|
|
2BFC
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
1SQG
 
 | | The crystal structure of the E. coli Fmu apoenzyme at 1.65 A resolution | | Descriptor: | SUN protein | | Authors: | Foster, P.G, Nunes, C.R, Greene, P, Moustakas, D, Stroud, R.M. | | Deposit date: | 2004-03-18 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The First Structure of an RNA m5C Methyltransferase,
Fmu, Provides Insight into Catalytic Mechanism
and Specific Binding of RNA Substrate
Structure, 11, 2003
|
|
4TZT
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS ENOYL REDUCTASE (INHA) COMPLEXED WITH N-(3-CHLORO-2-METHYLPHENYL)-1-CYCLOHEXYL- 5-OXOPYRROLIDINE-3-CARBOXAMIDE | | Descriptor: | (3S)-N-(3-CHLORO-2-METHYLPHENYL)-1-CYCLOHEXYL-5-OXOPYRROLIDINE-3-CARBOXAMIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, X, Alian, A, Stroud, R.M, Ortiz de Montellano, P.R. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis
J. Med. Chem., 2006
|
|
