2VO3
 
 | | Structure of PKA-PKB chimera complexed with C-(4-(4-Chlorophenyl)-1-(7H-pyrrolo(2,3-d)pyrimidin-4-yl)piperidin-4-yl)methylamine | | Descriptor: | 1-[4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-yl]methanamine, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Caldwell, J.J, Davies, T.G, Donald, A, McHardy, T, Rowlands, M.G, Aherne, G.W, Hunter, L.K, Taylor, K, Ruddle, R, Raynaud, F.I, Verdonk, M, Workman, P, Garrett, M.D, Collins, I. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Identification of 4-(4-Aminopiperidin-1-Yl)-7H-Pyrrolo[2,3-D]Pyrimidines as Selective Inhibitors of Protein Kinase B Through Fragment Elaboration.
J.Med.Chem., 51, 2008
|
|
2VUD
 
 | | PA-IIL lectin from pseudomonas aeruginosa complexed with fucose- derived glycomimetics | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, SULFATE ION, ... | | Authors: | Beha, S, Marotte, K, Sabin, C, Mitchell, E.P, Imberty, A, Roy, R. | | Deposit date: | 2008-05-22 | | Release date: | 2009-07-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fucose-Derived Glycomimetics as High Affinity Ligands for Bacterial Lectin Pa-Iil from Pseudomonas Aeruginosa
To be Published
|
|
2VPN
 
 | | High-resolution structure of the periplasmic ectoine-binding protein from TeaABC TRAP-transporter of Halomonas elongata | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, MAGNESIUM ION, PERIPLASMIC SUBSTRATE BINDING PROTEIN | | Authors: | Kuhlmann, S.I, Terwisscha van Scheltinga, A.C, Bienert, R, Kunte, H.J, Ziegler, C. | | Deposit date: | 2008-03-03 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 A Structure of the Ectoine Binding Protein Teaa of the Osmoregulated Trap-Transporter Teaabc from Halomonas Elongata.
Biochemistry, 47, 2008
|
|
2VOJ
 
 | | Ternary complex of M. tuberculosis Rv2780 with NAD and pyruvate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, ALANINE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2008-02-18 | | Release date: | 2008-03-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Mycobacterium Tuberculosis Secretory Antigen Alanine Dehydrogenase (Rv2780) in Apo and Ternary Complex Forms Captures "Open" and "Closed" Enzyme Conformations.
Proteins: Struct., Funct., Bioinf., 72, 2008
|
|
7BOI
 
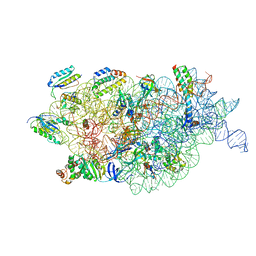 | | Bacterial 30S ribosomal subunit assembly complex state F (multibody refinement for body domain of 30S ribosome) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
2VQZ
 
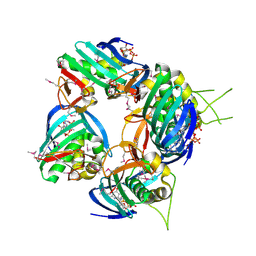 | | Structure of the cap-binding domain of influenza virus polymerase subunit PB2 with bound m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, POLYMERASE BASIC PROTEIN 2 | | Authors: | Guilligay, D, Tarendeau, F, Resa-Infante, P, Coloma, R, Crepin, T, Sehr, P, Lewis, J, Ruigrok, R.W.H, Ortin, J, Hart, D.J, Cusack, S. | | Deposit date: | 2008-03-21 | | Release date: | 2008-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structural Basis for CAP Binding by Influenza Virus Polymerase Subunit Pb2.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VUX
 
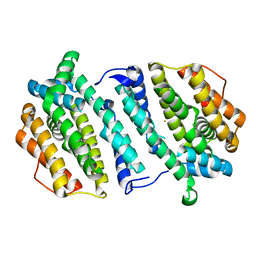 | | Human ribonucleotide reductase, subunit M2 B | | Descriptor: | FE (III) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE SUBUNIT M2 B | | Authors: | Welin, M, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Svensson, L, Thorsell, A.G, Tresaugues, L, van Den Berg, S, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-31 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human Ribonucleotide Reductase, Subunit M2 B
To be Published
|
|
7BOF
 
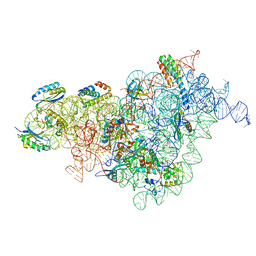 | | Bacterial 30S ribosomal subunit assembly complex state I (body domain) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7BJK
 
 | |
2W2F
 
 | | CRYSTAL STRUCTURE OF SINGLE POINT MUTANT ARG48GLN OF P-COUMARIC ACID DECARBOXYLASE FROM LACTOBACILLUS PLANTARUM STRUCTURAL INSIGHTS INTO THE ACTIVE SITE AND DECARBOXYLATION CATALYTIC MECHANISM | | Descriptor: | BARIUM ION, P-COUMARIC ACID DECARBOXYLASE | | Authors: | Rodriguez, H, Angulo, I, De Las Rivas, B, Campillo, N, Paez, J.A, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-10-29 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | P-Coumaric Acid Decarboxylase from Lactobacillus Plantarum: Structural Insights Into the Active Site and Decarboxylation Catalytic Mechanism.
Proteins, 78, 2010
|
|
2WBR
 
 | | The RRM domain in GW182 proteins contributes to miRNA-mediated gene silencing | | Descriptor: | GW182 | | Authors: | Eulalio, A, Tritschler, F, Buettner, R, Weichenrieder, O, Izaurralde, E, Truffault, V. | | Deposit date: | 2009-03-03 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Rrm Domain in Gw182 Proteins Contributes to Mirna-Mediated Gene Silencing.
Nucleic Acids Res., 37, 2009
|
|
2VO7
 
 | | Structure of PKA complexed with 4-(4-Chlorobenzyl)-1-(7H-pyrrolo(2,3- d)pyrimidin-4-yl)piperidin-4-ylamine | | Descriptor: | 4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-aminium, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Caldwell, J.J, Davies, T.G, Donald, A, McHardy, T, Rowlands, M.G, Aherne, G.W, Hunter, L.K, Taylor, K, Ruddle, R, Raynaud, F.I, Verdonk, M, Workman, P, Garrett, M.D, Collins, I. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Identification of 4-(4-Aminopiperidin-1-Yl)-7H-Pyrrolo[2,3-D]Pyrimidines as Selective Inhibitors of Protein Kinase B Through Fragment Elaboration.
J.Med.Chem., 51, 2008
|
|
7BMH
 
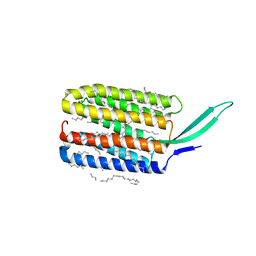 | | Crystal structure of a light-driven proton pump LR (Mac) from Leptosphaeria maculans | | Descriptor: | EICOSANE, OLEIC ACID, Opsin | | Authors: | Kovalev, K, Zabelskii, D, Dmitrieva, N, Volkov, O, Shevchenko, V, Astashkin, R, Zinovev, E, Gordeliy, V. | | Deposit date: | 2021-01-20 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based insights into evolution of rhodopsins.
Commun Biol, 4, 2021
|
|
7XH0
 
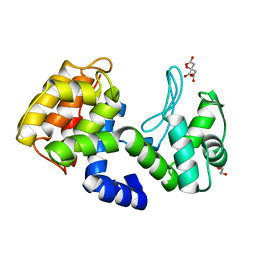 | | crystal structure of Csn-PD from Paenibacillus dendritiformis | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Chitosanase | | Authors: | Sun, H.H, Cheng, Y.M, Cao, R, Liu, Q, Zhao, L. | | Deposit date: | 2022-04-07 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | crystal structure of Csn-PD from Paenibacillus dendritiformis
To Be Published
|
|
7BOG
 
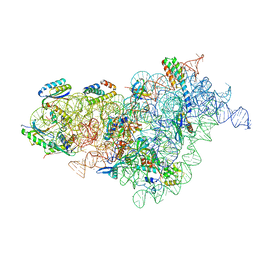 | | Bacterial 30S ribosomal subunit assembly complex state E (body domain) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
2VS0
 
 | |
2WDD
 
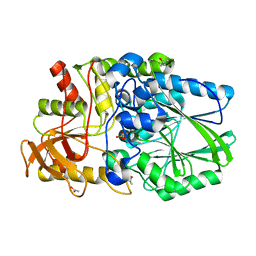 | | Termus thermophilus Sulfate thiohydrolase SoxB in complex with Sulfate | | Descriptor: | ACETATE ION, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Sauve, V, Roversi, P, Leath, K.J, Garman, E.F, Antrobus, R, Lea, S.M, Berks, B.C. | | Deposit date: | 2009-03-24 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism for the Hydrolysis of a Sulfur-Sulfur Bond Based on the Crystal Structure of the Thiosulfohydrolase Soxb.
J.Biol.Chem., 284, 2009
|
|
7BOD
 
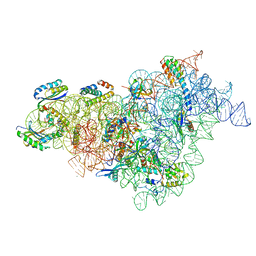 | | Bacterial 30S ribosomal subunit assembly complex state M (body domain) | | Descriptor: | 16S rRNA (body domain of 30S subunit), 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S.R. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7BF1
 
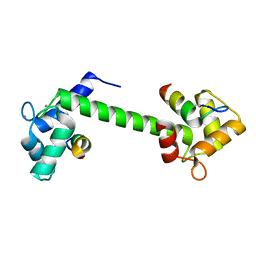 | | Ca2+-Calmodulin in complex with peptide from brain-type creatine kinase in extended 1:2 binding mode | | Descriptor: | ACETYL GROUP, CALCIUM ION, Calmodulin-1, ... | | Authors: | Sprenger, J, Akerfeldt, K.S, Bredfelt, J, Patel, N, Rowlett, R, Trifan, A, Vanderbeck, A, Lo Leggio, L, Snogerup Linse, S. | | Deposit date: | 2020-12-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Calmodulin complexes with brain and muscle creatine kinase peptides.
Curr Res Struct Biol, 3, 2021
|
|
2W1P
 
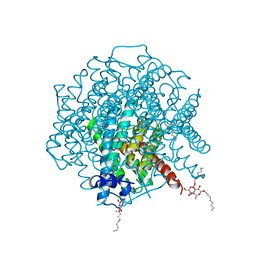 | | 1.4 Angstrom crystal structure of P.pastoris aquaporin, Aqy1, in a closed conformation at pH 8.0 | | Descriptor: | AQUAPORIN PIP2-7 7;, CHLORIDE ION, octyl beta-D-glucopyranoside | | Authors: | Fischer, G, Kosinska-Eriksson, U, Aponte-Santamaria, C, Palmgren, M, Geijer, C, Hedfalk, K, Hohmann, S, de Groot, B.L, Neutze, R, Lindkvist-Petersson, K. | | Deposit date: | 2008-10-20 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Yeast Aquaporin at 1.15 A Reveals a Novel Gating Mechanism
Plos Biol., 7, 2009
|
|
7BF2
 
 | | Ca2+-Calmodulin in complex with human muscle form creatine kinase peptide in extended 1:2 binding mode | | Descriptor: | CALCIUM ION, Calmodulin-1, Creatine kinase M-type | | Authors: | Sprenger, J, Akerfeldt, K.S, Bredfelt, J, Patel, N, Rowlett, R, Trifan, A, Vanderbeck, A, Lo Leggio, L, Snogerup Linse, S. | | Deposit date: | 2020-12-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Calmodulin complexes with brain and muscle creatine kinase peptides.
Curr Res Struct Biol, 3, 2021
|
|
2VZV
 
 | | Substrate Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with chitosan | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, EXO-BETA-D-GLUCOSAMINIDASE | | Authors: | Lammerts van Bueren, A, Ghinet, M.G, Gregg, K, Fleury, A, Brzezinski, R, Boraston, A.B. | | Deposit date: | 2008-08-05 | | Release date: | 2008-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structural Basis of Substrate Recognition in an Exo-Beta-D-Glucosaminidase Involved in Chitosan Hydrolysis.
J.Mol.Biol., 385, 2009
|
|
3JUO
 
 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with (R)-5-bromo-2-(piperidin-3-ylamino)benzoic acid | | Descriptor: | 5-bromo-2-[(3R)-piperidin-3-ylamino]benzoic acid, Phenazine biosynthesis protein A/B | | Authors: | Mentel, M, Jain, I.H, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site of an Enzyme Can Host Both Enantiomers of a Racemic Ligand Simultaneously
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2W2G
 
 | | Human SARS coronavirus unique domain | | Descriptor: | NON-STRUCTURAL PROTEIN 3, SULFATE ION | | Authors: | Tan, J, Vonrhein, C, Smart, O.S, Bricogne, G, Bollati, M, Kusov, Y, Hansen, G, Mesters, J.R, Schmidt, C.L, Hilgenfeld, R. | | Deposit date: | 2008-10-30 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The Sars-Unique Domain (Sud) of Sars Coronavirus Contains Two Macrodomains that Bind G-Quadruplexes.
Plos Pathog., 5, 2009
|
|
2W46
 
 | | CBM35 from Cellvibrio japonicus Abf62 | | Descriptor: | CALCIUM ION, ESTERASE D, SODIUM ION | | Authors: | Montainer, C, Lammerts van Bueren, A, Dumon, C, Flint, J.E, Correia, M.A, Prates, J.A, Firbank, S.J, Lewis, R.J, Grondin, G.G, Ghinet, M.G, Gloster, T.M, Herve, C, Knox, J.P, Talbot, B.G, Turkenburg, J.P, Kerovuo, J, Brzezinski, R, Fontes, C.M.G.A, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-01-27 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
