2WW5
 
 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae at 1.6 A resolution | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L, Menendez, M. | | Deposit date: | 2009-10-21 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7NT2
 
 | | Crystal structure of SARS CoV2 main protease in complex with FSP006 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6ZG1
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
7OQH
 
 | | CryoEM structure of the transcription termination factor Rho from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Transcription termination factor Rho | | Authors: | Saridakis, E, Vishwakarma, R, Lai Kee Him, J, Martin, K, Simon, I, Cohen-Gonsaud, M, Coste, F, Bron, P, Margeat, E, Boudvillain, M. | | Deposit date: | 2021-06-03 | | Release date: | 2022-02-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structure of transcription termination factor Rho from Mycobacterium tuberculosis reveals bicyclomycin resistance mechanism.
Commun Biol, 5, 2022
|
|
5YYM
 
 | | Crystal structures of E.coli arginyl-trna synthetase (argrs) in complex with substrate Arg | | Descriptor: | ARGININE, Arginine--tRNA ligase | | Authors: | Zhou, M, Ye, S, Stephen, P, Zhang, R.G, Wang, E.D, Giege, R, Lin, S.X. | | Deposit date: | 2017-12-10 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures Of E.Coli Arginyl-Trna Synthetase (Argrs) In Complex With Substrates
To Be Published
|
|
3GGL
 
 | | X-Ray Structure of the C-terminal domain (277-440) of Putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR324A. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative chitobiase, ZINC ION | | Authors: | Kuzin, A, Neely, H, Seetharaman, R, Lee, D, Ciccosanti, C, Foote, E.L, Janjua, H, Xiao, R, Nair, R, Rost, B, Acton, T, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-28 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Northeast Structural Genomics Consortium Target BtR324A
To be Published
|
|
4KXV
 
 | | Human transketolase in covalent complex with donor ketose D-xylulose-5-phosphate, crystal 1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, D-XYLITOL-5-PHOSPHATE, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
6Z6F
 
 | | HDAC-PC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
5Z7O
 
 | | SmChiA sliding-intermediate with chitotetraose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A, GLYCEROL | | Authors: | Nakamura, A, Iino, R. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Processive chitinase is Brownian monorail operated by fast catalysis after peeling rail from crystalline chitin.
Nat Commun, 9, 2018
|
|
6Z6H
 
 | | HDAC-DC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (8.55 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
7P6G
 
 | | Crystal structure of the endoglucanase RBcel1 E135Q | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase | | Authors: | Collet, L, Dutoit, R. | | Deposit date: | 2021-07-16 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Highlighting the factors governing transglycosylation in the GH5_5 endo-1,4-beta-glucanase RBcel1.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6Z6O
 
 | | HDAC-TC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
7P6J
 
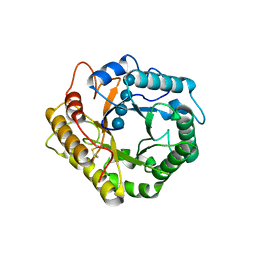 | | Crystal structure of glycosyl-enzyme intermediate of RBcel1 Y201F | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Collet, L, Dutoit, R. | | Deposit date: | 2021-07-16 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Highlighting the factors governing transglycosylation in the GH5_5 endo-1,4-beta-glucanase RBcel1.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
2OB7
 
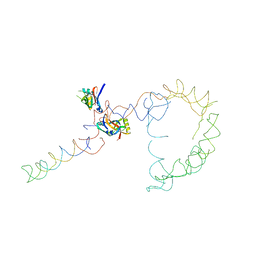 | | Structure of tmRNA-(SmpB)2 complex as inferred from cryo-EM | | Descriptor: | 16S ribosomal RNA, SsrA-binding protein, transfer-messenger RNA | | Authors: | Frank, J, Felden, B, Gillet, R, Li, W. | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | Scaffolding as an organizing principle in trans-translation. The roles of small protein B and ribosomal protein S1.
J.Biol.Chem., 282, 2007
|
|
7P6I
 
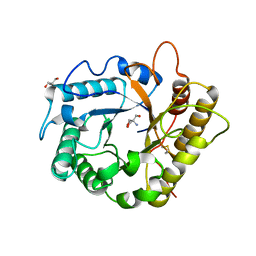 | | Crystal structure of the endoglucanase RBcel1 Y201F | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase | | Authors: | Collet, L, Dutoit, R. | | Deposit date: | 2021-07-16 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Highlighting the factors governing transglycosylation in the GH5_5 endo-1,4-beta-glucanase RBcel1.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7P6H
 
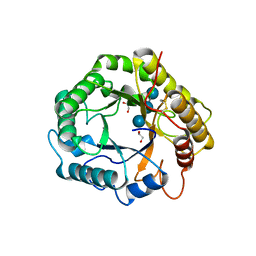 | |
6Z6P
 
 | | HDAC-PC-Nuc | | Descriptor: | DNA (145-MER), HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
5YU6
 
 | | CRYSTAL STRUCTURE OF EXPORTIN-5:RANGTP COMPLEX | | Descriptor: | 13-mer peptide, Exportin-5, GTP-binding nuclear protein Ran, ... | | Authors: | Yamazawa, R, Jiko, C, Lee, S.J, Yamashita, E. | | Deposit date: | 2017-11-20 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural Basis for Selective Binding of Export Cargoes by Exportin-5
Structure, 26, 2018
|
|
6Z1C
 
 | | Crystal structure of Arabidopsis thaliana CK2-alpha-1 in complex with TTP-22 | | Descriptor: | 3-[5-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]sulfanylpropanoic acid, CHLORIDE ION, Casein kinase II subunit alpha-1 | | Authors: | Demulder, M, Loris, R, De Veylder, L. | | Deposit date: | 2020-05-13 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arabidopsis casein kinase 2 triggers stem cell exhaustion under Al toxicity and phosphate deficiency through activating the DNA damage response pathway.
Plant Cell, 33, 2021
|
|
6TNJ
 
 | |
7P7G
 
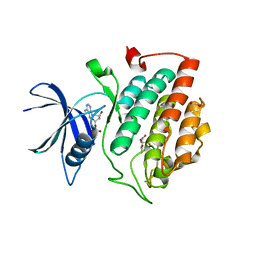 | | Crystal structure of phosphorylated pT220 Casein Kinase I delta (CK1d), conformation 2 and 3 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CITRIC ACID, ... | | Authors: | Chaikuad, A, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinase domain autophosphorylation rewires the activity and substrate specificity of CK1 enzymes.
Mol.Cell, 82, 2022
|
|
7P7H
 
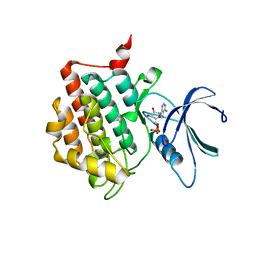 | |
7P7F
 
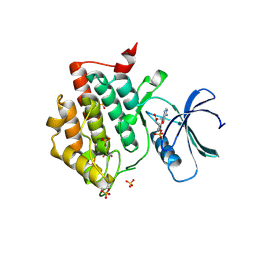 | | Crystal structure of phosphorylated pT220 Casein Kinase I delta (CK1d), conformation 1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Chaikuad, A, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Kinase domain autophosphorylation rewires the activity and substrate specificity of CK1 enzymes.
Mol.Cell, 82, 2022
|
|
3DPE
 
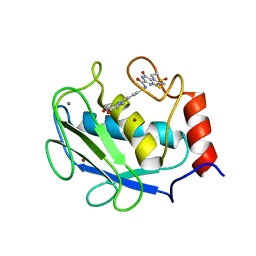 | | Crystal structure of the complex between MMP-8 and a non-zinc chelating inhibitor | | Descriptor: | CALCIUM ION, N-{[2-(2-amino-3,4-dioxocyclobut-1-en-1-yl)-1,2,3,4-tetrahydroisoquinolin-7-yl]methyl}-4-oxo-3,5,6,8-tetrahydro-4H-thiopyrano[4',3':4,5]thieno[2,3-d]pyrimidine-2-carboxamide 7,7-dioxide, Neutrophil collagenase, ... | | Authors: | Pochetti, G, Montanari, R, Mazza, F. | | Deposit date: | 2008-07-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Extra Binding Region Induced by Non-Zinc Chelating Inhibitors into the S(1)' Subsite of Matrix Metalloproteinase 8 (MMP-8)
J.Med.Chem., 52, 2009
|
|
5YYN
 
 | | Crystal structures of E.coli arginyl-trna synthetase (argrs) in complex with substrate TRNA(Arg) | | Descriptor: | Arginine--tRNA ligase, TRNA | | Authors: | Zhou, M, Ye, S, Stephen, P, Zhang, R.G, Wang, E.D, Giege, R, Lin, S.X. | | Deposit date: | 2017-12-10 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures Of E.Coli Arginyl-Trna Synthetase (Argrs) In Complex With Substrates
To Be Published
|
|
