2X39
 
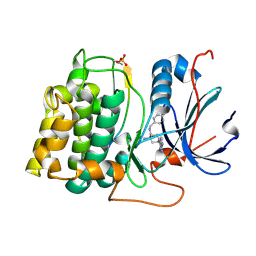 | | Structure of 4-Amino-N-(4-chlorobenzyl)-1-(7H-pyrrolo(2,3-d)pyrimidin- 4-yl)piperidine-4-carboxamide bound to PKB | | 分子名称: | 4-AMINO-N-(4-CHLOROBENZYL)-1-(7H-PYRROLO[2,3-D]PYRIMIDIN-4-YL)PIPERIDINE-4-CARBOXAMIDE, GLYCOGEN SYNTHASE KINASE-3 BETA, RAC-BETA SERINE/THREONINE-PROTEIN KINASE | | 著者 | Davies, T.G, McHardy, T, Caldwell, J.J, Cheung, K.M, Hunter, L.J, Taylor, K, Rowlands, M, Ruddle, R, Henley, A, Brandon, A.D, Valenti, M, Fazal, L, Seavers, L, Raynaud, F.I, Eccles, S.A, Aherne, G.W, Garrett, M.D, Collins, I. | | 登録日 | 2010-01-22 | | 公開日 | 2010-02-23 | | 最終更新日 | 2011-09-21 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Discovery of 4-Amino-1-(7H-Pyrrolo[2,3-D]Pyrimidin-4-Yl)Piperidine-4-Carboxamides as Selective, Orally Active Inhibitors of Protein Kinase B (Akt).
J.Med.Chem., 53, 2010
|
|
7BCV
 
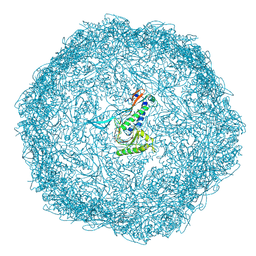 | |
3JPV
 
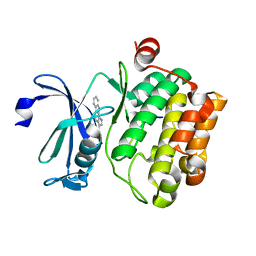 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a pyrrolo[2,3-a]carbazole ligand | | 分子名称: | 1,10-dihydropyrrolo[2,3-a]carbazole-3-carbaldehyde, Peptide (PIMTIDE) ARKRRRHPSGPPTA, Proto-oncogene serine/threonine-protein kinase Pim-1 | | 著者 | Filippakopoulos, P, Bullock, A.N, Fedorov, O, Akue-Gedu, R, Rossignol, E, Azzaro, S, Bain, J, Cohen, P, Prudhomme, M, Moreau, P, Amizon, F, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2009-09-04 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Synthesis, kinase inhibitory potencies, and in vitro antiproliferative evaluation of new pim kinase inhibitors.
J.Med.Chem., 52, 2009
|
|
7BQE
 
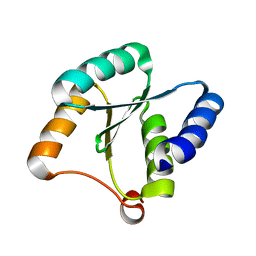 | | Solution NMR structure of NF3; de novo designed protein with a novel fold | | 分子名称: | NF3 | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
2X9W
 
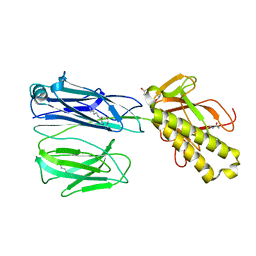 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | 分子名称: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, GLYCEROL | | 著者 | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | 登録日 | 2010-03-25 | | 公開日 | 2010-06-30 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
7C1M
 
 | | Complex structure of tyrosinated alpha-tubulin carboxy-terminal peptide and A1aY1 binder | | 分子名称: | Carboxy-terminal peptide from tyrosinated alpha-tubulin, Nanobody binder from SSO7d library | | 著者 | Kesarwani, S, Reddy, P.P, Sirajuddin, M, Das, R. | | 登録日 | 2020-05-05 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Genetically encoded live-cell sensor for tyrosinated microtubules.
J.Cell Biol., 219, 2020
|
|
7BQQ
 
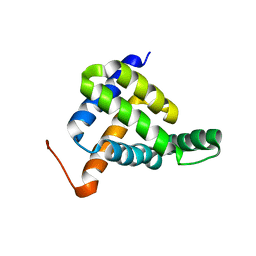 | | Solution NMR structure of fold-Z Gogy; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Gogy | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQM
 
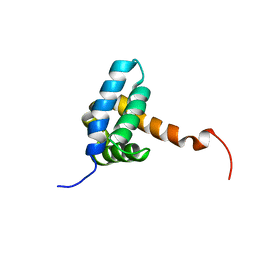 | | Solution NMR structure of fold-0 Chantal; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Chantal | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BBB
 
 | |
7BQB
 
 | | Solution NMR structure of NF6; de novo designed protein with a novel fold | | 分子名称: | NF6 | | 著者 | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQC
 
 | | Solution NMR structure of NF4; de novo designed protein with a novel fold | | 分子名称: | NF4 | | 著者 | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, T, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQN
 
 | | Solution NMR structure of fold-C Rei; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Rei | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7C4Z
 
 | | Cryo-EM structure of empty Coxsackievirus A10 at pH 5.5 | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | 著者 | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | 登録日 | 2020-05-18 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3JUM
 
 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with 5-bromo-2-((1S,3R)-3-carboxycyclohexylamino)benzoic acid | | 分子名称: | 5-bromo-2-{[(1S,3R)-3-carboxycyclohexyl]amino}benzoic acid, Phenazine biosynthesis protein A/B | | 著者 | Mentel, M, Breinbauer, R, Blankenfeldt, W. | | 登録日 | 2009-09-15 | | 公開日 | 2009-09-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The Active Site of an Enzyme Can Host Both Enantiomers of a Racemic Ligand Simultaneously
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
7C6F
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in an open state | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | 登録日 | 2020-05-21 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7C6M
 
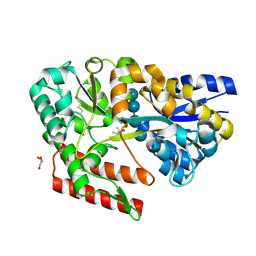 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to cellotetraose (Form I) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | 登録日 | 2020-05-21 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
2WT8
 
 | |
7C6Y
 
 | | Crystal structure of beta-glycosides-binding protein (W41A) of ABC transporter in an open state (Form II) | | 分子名称: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CHLORIDE ION, ... | | 著者 | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | 登録日 | 2020-05-22 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
2WU3
 
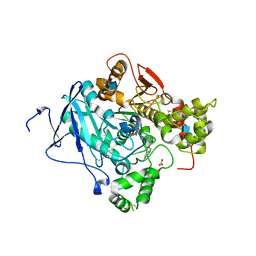 | | CRYSTAL STRUCTURE OF MOUSE ACETYLCHOLINESTERASE IN COMPLEX WITH FENAMIPHOS AND HI-6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ACETYLCHOLINESTERASE, ... | | 著者 | Hornberg, A, Artursson, E, Warme, R, Pang, Y.-P, Ekstrom, F. | | 登録日 | 2009-09-28 | | 公開日 | 2009-10-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structures of Oxime-Bound Fenamiphos-Acetylcholinesterases: Reactivation Involving Flipping of the His447 Ring to Form a Reactive Glu334-His447-Oxime Triad.
Biochem.Pharm., 79, 2010
|
|
7C67
 
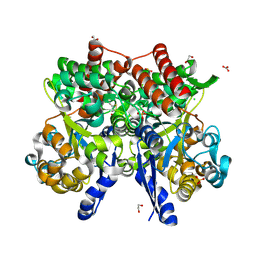 | | Crystal structure of beta-glycosides-binding protein of ABC transporter in a closed state bound to cellotriose | | 分子名称: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | 著者 | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | 登録日 | 2020-05-21 | | 公開日 | 2020-10-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7X8L
 
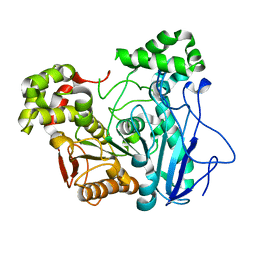 | |
7B53
 
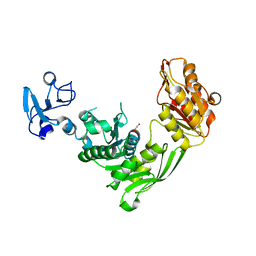 | | Crystal structure of MurE from E.coli | | 分子名称: | 1,2-ETHANEDIOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase | | 著者 | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | 登録日 | 2020-12-03 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of MurE from E.coli
To Be Published
|
|
2XAF
 
 | | Crystal structure of LSD1-CoREST in complex with para-bromo-(+)-cis-2- phenylcyclopropyl-1-amine | | 分子名称: | 3-(4-BROMOPHENYL)PROPANAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | 著者 | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | 登録日 | 2010-03-31 | | 公開日 | 2010-05-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
1AQE
 
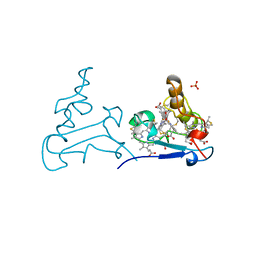 | |
7AV9
 
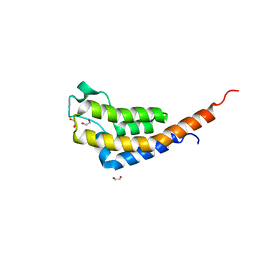 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 | | 分子名称: | 1,2-ETHANEDIOL, PH-interacting protein | | 著者 | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | 登録日 | 2020-11-04 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2
To Be Published
|
|
