6TTO
 
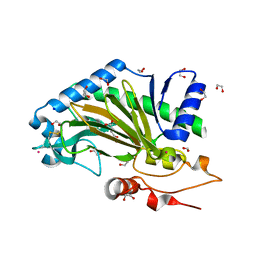 | | N-terminally truncated hyoscyamine 6-hydroxylase (tH6H) in complex with 2-oxoglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FORMIC ACID, ... | | Authors: | Kluza, A, Mrugala, B, Porebski, P.J, Kurpiewska, K, Niedzialkowska, E, Weiss, M.S, Borowski, T. | | Deposit date: | 2019-12-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Regioselectivity of hyoscyamine 6 beta-hydroxylase-catalysed hydroxylation as revealed by high-resolution structural information and QM/MM calculations.
Dalton Trans, 49, 2020
|
|
6S0U
 
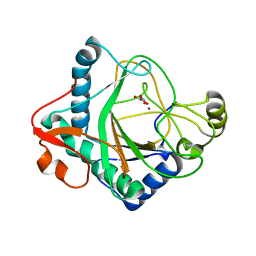 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6S0T
 
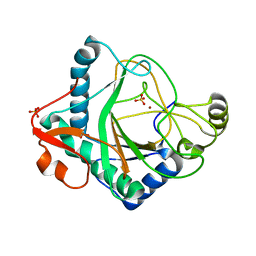 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel, sulfate, soaked with iodide | | Descriptor: | IODIDE ION, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | Authors: | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Cymborowski, M.T, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6CHK
 
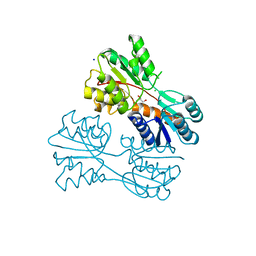 | | Crystal structure of LacI family transcriptional regulator from Lactobacillus casei, Target EFI-512911, with bound TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Shabalin, I.G, Kowiel, M, Porebski, P.J, Minor, W, Jaskolski, M, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative, E.F.I. | | Deposit date: | 2018-02-22 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Automatic recognition of ligands in electron density by machine learning.
Bioinformatics, 35, 2019
|
|
3MLE
 
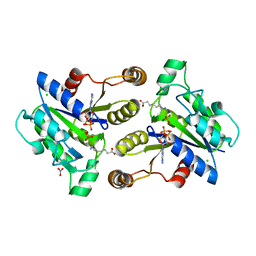 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori cocrystallized with ATP | | Descriptor: | 8-aminooctanoic acid, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Nicholls, R, Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-16 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
6S0R
 
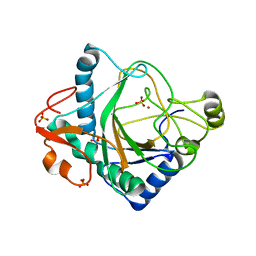 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus complex with nickel, sulfate and chloride | | Descriptor: | CHLORIDE ION, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | Authors: | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Cymborowski, M.T, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
5C5I
 
 | | Crystal structure of NADP-dependent dehydrogenase from Rhodobacter sphaeroides | | Descriptor: | NADP-dependent dehydrogenase | | Authors: | Kowiel, M, Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Porebski, P.J, Cymborowski, M, Al Obaidi, N.F, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-19 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of NADP-dependent dehydrogenase from Rhodobacter sphaeroides
to be published
|
|
3UWD
 
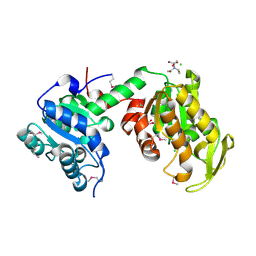 | | Crystal Structure of Phosphoglycerate Kinase from Bacillus Anthracis | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zheng, H, Chruszcz, M, Porebski, P, Kudritska, M, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-01 | | Release date: | 2012-01-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structures of putative phosphoglycerate kinases from B. anthracis and C. jejuni.
J.Struct.Funct.Genom., 13, 2012
|
|
6TTM
 
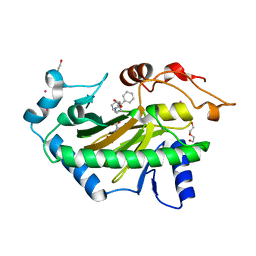 | | Hyoscyamine 6-hydroxylase in complex with N-oxalylglycine and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Hyoscyamine 6 beta-hydroxylase, ... | | Authors: | Kluza, A, Kurpiewska, K, Mrugala, B, Porebski, P.J, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-12-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Regioselectivity of hyoscyamine 6 beta-hydroxylase-catalysed hydroxylation as revealed by high-resolution structural information and QM/MM calculations.
Dalton Trans, 49, 2020
|
|
6TTN
 
 | | N-terminally truncated hyoscyamine 6-hydroxylase (tH6H) in complex with N-oxalylglycine and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, Hyoscyamine 6 beta-hydroxylase, N-OXALYLGLYCINE, ... | | Authors: | Kluza, A, Mrugala, B, Porebski, P.J, Kurpiewska, K, Niedzialkowska, E, Weiss, M.S, Borowski, T. | | Deposit date: | 2019-12-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Regioselectivity of hyoscyamine 6 beta-hydroxylase-catalysed hydroxylation as revealed by high-resolution structural information and QM/MM calculations.
Dalton Trans, 49, 2020
|
|
6CIG
 
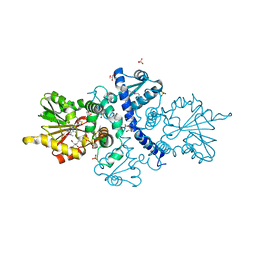 | | CRYSTAL STRUCTURE ANALYSIS OF SELENOMETHIONINE SUBSTITUTED ISOFLAVONE O-METHYLTRANSFERASE | | Descriptor: | GLYCEROL, Isoflavone-7-O-methyltransferase 8, N-(TRIS(HYDROXYMETHYL)METHYL)-3-AMINOPROPANESULFONIC ACID, ... | | Authors: | Zubieta, C, Dixon, R.A, Shabalin, I.G, Kowiel, M, Porebski, P.J, Noel, J.P. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-07 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases.
Nat. Struct. Biol., 8, 2001
|
|
3N0S
 
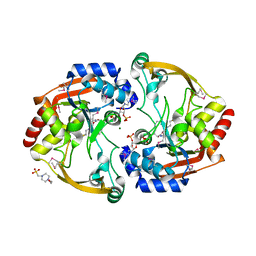 | | Crystal structure of BA2930 mutant (H183A) in complex with AcCoA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-14 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
3N0M
 
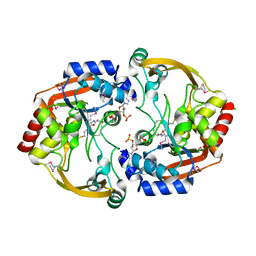 | | Crystal structure of BA2930 mutant (H183G) in complex with AcCoA | | Descriptor: | ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, CHLORIDE ION | | Authors: | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-14 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
3OT1
 
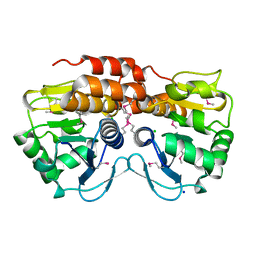 | | Crystal structure of VC2308 protein | | Descriptor: | 4-methyl-5(B-hydroxyethyl)-thiazole monophosphate biosynthesis enzyme, CHLORIDE ION, SODIUM ION | | Authors: | Niedzialkowska, E, Wawrzak, Z, Chruszcz, M, Porebski, P, Skarina, T, Huang, X, Grimshaw, S, Cymborowski, M, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-10 | | Release date: | 2010-09-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of VC2308 protein
To be Published
|
|
5VET
 
 | | PHOSPHOLIPASE A2, RE-REFINEMENT OF THE PDB STRUCTURE 1JQ8 WITHOUT THE PUTATIVE COMPLEXED OLIGOPEPTIDE | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2018-06-20 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VER
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Z | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEP
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2F | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEQ
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Y | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEH
 
 | | Re-refinement OF THE PDB STRUCTURE 1yiz of Aedes aegypti kynurenine aminotransferase | | Descriptor: | BROMIDE ION, GLYCEROL, Kynurenine aminotransferase | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
4QGL
 
 | | Acireductone dioxygenase from Bacillus anthracis with three cadmium ions | | Descriptor: | Acireductone dioxygenase, CADMIUM ION | | Authors: | Milaczewska, A.M, Chruszcz, M, Majorek, K.A, Porebski, P.J, Borowski, T, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-05-23 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Acireductone dioxygenase from Bacillus anthracis with three cadmium ions
To be Published
|
|
6CNY
 
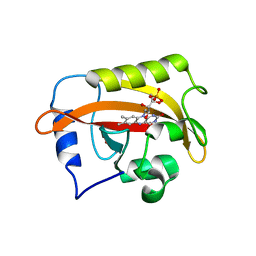 | | 2.3 Angstrom Structure of Phosphodiesterase treated Vivid (complex with FMN) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Vivid PAS protein VVD | | Authors: | Zoltowski, B.D, Shabalin, I.G, Kowiel, M, Porebski, P.J, Crane, B.R, Bilwes, A.M. | | Deposit date: | 2018-03-09 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational switching in the fungal light sensor Vivid.
Science, 316, 2007
|
|
3UED
 
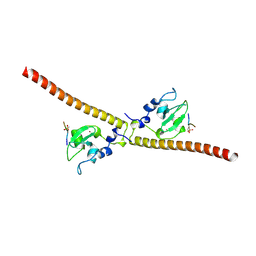 | | Crystal structure of human Survivin bound to histone H3 phosphorylated on threonine-3 (C2 space group). | | Descriptor: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEH
 
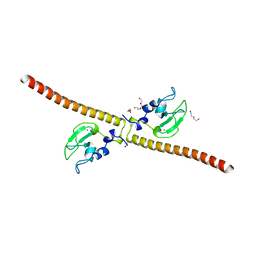 | | Crystal structure of human Survivin H80A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3UEE
 
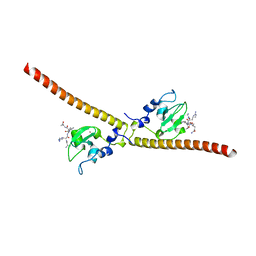 | | Crystal structure of human Survivin K62A mutant bound to N-terminal histone H3 | | Descriptor: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3V09
 
 | | Crystal structure of Rabbit Serum Albumin | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
