2LSM
 
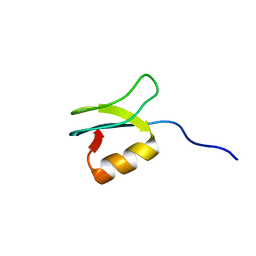 | | Solution structure of gpFI C-terminal domain | | Descriptor: | DNA-packaging protein FI | | Authors: | Popovic, A, Wu, B, Edwards, A.M, Davidson, A.R, Maxwell, K.L. | | Deposit date: | 2012-05-02 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and biochemical characterization of phage lambda FI protein (gpFI) reveals a novel mechanism of DNA packaging chaperone activity.
J.Biol.Chem., 287, 2012
|
|
1BZ6
 
 | |
5D8M
 
 | | Crystal structure of the metagenomic carboxyl esterase MGS0156 | | Descriptor: | Metagenomic carboxyl esterase MGS0156 | | Authors: | Cui, H, Nocek, B, Tchigvintsev, A, Popovic, A, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2015-08-17 | | Release date: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of esterase (MGS0156)
To Be Published
|
|
4JHC
 
 | | Crystal structure of the uncharacterized Maf protein YceF from E. coli | | Descriptor: | GLYCEROL, Maf-like protein YceF, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Xu, X, Cui, H, Tchigvintsev, A, Flick, R, Brown, G, Popovic, A, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2013-03-04 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural studies of conserved maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides.
Chem.Biol., 20, 2013
|
|
4LU1
 
 | | Crystal structure of the uncharacterized Maf protein YceF from E. coli, mutant D69A | | Descriptor: | Maf-like protein YceF, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Xu, X, Cui, H, Tchigvintsev, A, Flick, R, Brown, G, Popovic, A, Yakunin, A.F, Savchenko, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and structural studies of conserved maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides.
Chem.Biol., 20, 2013
|
|
4V7G
 
 | | Crystal Structure of Lumazine Synthase from Bacillus Anthracis | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Morgunova, E, Illarionov, B, Saller, S, Popov, A, Sambaiah, T, Bacher, A, Cushman, M, Fischer, M, Ladenstein, R. | | Deposit date: | 2009-09-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural study and thermodynamic characterization of inhibitor binding to lumazine synthase from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2AUU
 
 | | Inorganic pyrophosphatase complexed with magnesium pyrophosphate and fluoride | | Descriptor: | CHLORIDE ION, FLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Samygina, V.R, Popov, A.N, Avaeva, S.M, Bartunik, H.D. | | Deposit date: | 2005-08-29 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Reversible inhibition of Escherichia coli inorganic pyrophosphatase by fluoride: trapped catalytic intermediates in cryo-crystallographic studies
J.Mol.Biol., 366, 2007
|
|
8RV0
 
 | | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in complex with nitrite | | Descriptor: | CALCIUM ION, HEME C, NITRIC OXIDE, ... | | Authors: | Polyakov, K.M, Safonova, T.N, Osipov, E, Popov, A.N, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-01-31 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in complex with nitrite
To Be Published
|
|
8RXU
 
 | | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in space group P21 | | Descriptor: | CALCIUM ION, HEME C, Octaheme nitrite reductase, ... | | Authors: | Polyakov, K.M, Safonova, T.N, Osipov, E, Popov, A.N, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-02-08 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in space group P21
To Be Published
|
|
8RVM
 
 | | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in space group P63 | | Descriptor: | CALCIUM ION, HEME C, Octaheme nitrite c cytochrome c reductase, ... | | Authors: | Polyakov, K.M, Safoonova, T.N, Osipov, E, Popov, A.N, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-02-01 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in space group P63
To Be Published
|
|
2ZO5
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with azide | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-05-05 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens
J.Mol.Biol., 389, 2009
|
|
6EIG
 
 | | Crystal structure of N24Q/C128T mutant of Channelrhodopsin 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Archaeal-type opsin 2, EICOSANE, ... | | Authors: | Kovalev, K, Borshchevskiy, V, Volkov, O, Polovinkin, V, Marin, E, Balandin, T, Astashkin, R, Bamann, C, Bueldt, G, Willlbold, D, Popov, A, Bamberg, E, Gordeliy, V. | | Deposit date: | 2017-09-19 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into ion conduction by channelrhodopsin 2.
Science, 358, 2017
|
|
6EID
 
 | | Crystal structure of wild-type Channelrhodopsin 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Archaeal-type opsin 2, PHOSPHATE ION, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Volkov, O, Polovinkin, V, Marin, E, Balandin, T, Astashkin, R, Bamann, C, Bueldt, G, Willlbold, D, Popov, A, Bamberg, E, Gordeliy, V. | | Deposit date: | 2017-09-19 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights into ion conduction by channelrhodopsin 2.
Science, 358, 2017
|
|
6ES3
 
 | | Structure of CDX2-DNA(TCG) | | Descriptor: | DNA (5'-D(P*GP*GP*AP*GP*GP*TP*CP*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*CP*GP*AP*CP*CP*TP*CP*C)-3'), Homeobox protein CDX-2 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
6ES2
 
 | | Structure of CDX2-DNA(CAA) | | Descriptor: | DNA (5'-D(P*GP*GP*AP*GP*GP*CP*AP*AP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*TP*TP*GP*CP*CP*TP*CP*C)-3'), Homeobox protein CDX-2 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
6I3Q
 
 | | The structure of thiocyanate dehydrogenase from Thioalkalivibrio paradoxus complex with acetate ions. | | Descriptor: | ACETATE ION, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Polyakov, K.M, Popov, A.N, Tikhkonova, T.V, Popov, V.O, Trofimov, A.A. | | Deposit date: | 2018-11-07 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8BYX
 
 | |
1VPI
 
 | | PHOSPHOLIPASE A2 INHIBITOR FROM VIPOXIN | | Descriptor: | PHOSPHOLIPASE A2 INHIBITOR | | Authors: | Devedjiev, Y.D, Popov, A.N. | | Deposit date: | 1996-12-17 | | Release date: | 1997-12-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray structure at 1.76 A resolution of a polypeptide phospholipase A2 inhibitor.
J.Mol.Biol., 266, 1997
|
|
8BZM
 
 | | FOXK1-ELF1-heterodimer bound to DNA | | Descriptor: | DNA, ETS-related transcription factor Elf-1, Forkhead box protein K1, ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | FOXK1-ELF1_heterodimer bound to DNA
To Be Published
|
|
3D1I
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with nitrite | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-05-06 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens
J.Mol.Biol., 389, 2009
|
|
1YA5
 
 | | Crystal structure of the titin domains z1z2 in complex with telethonin | | Descriptor: | N2B-TITIN ISOFORM, SULFATE ION, TELETHONIN | | Authors: | Pinotsis, N, Popov, A, Zou, P, Wilmanns, M. | | Deposit date: | 2004-12-17 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Palindromic assembly of the giant muscle protein titin in the sarcomeric Z-disk
Nature, 439, 2006
|
|
5TJE
 
 | | Murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 and T cell receptor P14 | | Descriptor: | ALPHA CHAIN OF MURINE T CELL RECEPTOR p14, BETA CHAIN OF MURINE T CELL RECEPTOR p14, Beta-2-microglobulin, ... | | Authors: | Achour, A, Sandalova, T, Allerbring, E, Popov, A. | | Deposit date: | 2016-10-04 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
to be published
|
|
2V8T
 
 | | Crystal structure of Mn catalase from Thermus Thermophilus complexed with chloride | | Descriptor: | CHLORIDE ION, LITHIUM ION, MANGANESE (II) ION, ... | | Authors: | Antonyuk, S.V, Barynin, V.V, Vaguine, A.A, Melik-Adamyan, W.R, Popov, A.N, Lamsin, V.S, Harrison, P.M, Artymiuk, P.J. | | Deposit date: | 2007-08-14 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Three-Dimentional Structure of the Enzyme Dimanganese Catalase from Thermus Thermophilus at 1 Angstrom Resolution
Crystallogr.Rep.(Transl. Kristallografiya), 45, 2000
|
|
2V8U
 
 | | Atomic resolution structure of Mn catalase from Thermus Thermophilus | | Descriptor: | LITHIUM ION, MANGANESE (II) ION, MANGANESE-CONTAINING PSEUDOCATALASE, ... | | Authors: | Barynin, V.V, Antonyuk, S.V, Vaguine, A.A, Melik-Adamyan, W.R, Popov, A.N, Lamsin, V.S, Harrison, P.M, Artymiuk, P.J. | | Deposit date: | 2007-08-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Three-Dimentional Structure of the Enzyme Dimanganese Catalase from Thermus Thermophilus at 1 Angstrom Resolution
Crystallogr.Rep.(Transl. Kristallografiya), 45, 2000
|
|
7Q37
 
 | | Crystal structure of proton pump MAR rhodopsin pressurized with krypton | | Descriptor: | Bacteriorhodopsin, EICOSANE, HEXANE, ... | | Authors: | Melnikov, I, Rulev, M, Astashkin, R, Kovalev, K, Carpentier, P, Gordeliy, V, Popov, A. | | Deposit date: | 2021-10-27 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-pressure crystallography shows noble gas intervention into protein-lipid interaction and suggests a model for anaesthetic action.
Commun Biol, 5, 2022
|
|
