2GO1
 
 | | NAD-dependent formate dehydrogenase from Pseudomonas sp.101 | | 分子名称: | NAD-dependent formate dehydrogenase, SULFATE ION | | 著者 | Filippova, E.V, Polyakov, K.M, Tikhonova, T.V, Stekhanova, T.N, Boiko, K.M, Popov, V.O. | | 登録日 | 2006-04-12 | | 公開日 | 2006-05-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of a new crystal modification of the bacterial NAD-dependent formate dehydrogenase with a resolution of 2.1 A
Crystallography reports, 50, 2005
|
|
2NDP
 
 | | Structure of DNA-binding HU protein from micoplasma Mycoplasma gallisepticum | | 分子名称: | Histone-like DNA-binding superfamily protein | | 著者 | Altukhov, D.A, Talyzina, A.A, Agapova, Y.K, Vlaskina, A.V, Korzhenevskiy, D.A, Bocharov, E.V, Rakitina, T.V, Timofeev, V.I, Popov, V.O. | | 登録日 | 2016-09-13 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Enhanced conformational flexibility of the histone-like (HU) protein from Mycoplasma gallisepticum.
J.Biomol.Struct.Dyn., 36, 2018
|
|
3FN4
 
 | | Apo-form of NAD-dependent formate dehydrogenase from bacterium Moraxella sp.C-1 in closed conformation | | 分子名称: | GLYCEROL, NAD-dependent formate dehydrogenase, SULFATE ION | | 著者 | Shabalin, I.G, Polyakov, K.M, Filippova, E.V, Dorovatovskiy, P.V, Tikhonova, T.V, Sadykhov, E.G, Tishkov, V.I, Popov, V.O. | | 登録日 | 2008-12-23 | | 公開日 | 2009-12-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structures of the apo and holo forms of formate dehydrogenase from the bacterium Moraxella sp. C-1: towards understanding the mechanism of the closure of the interdomain cleft
Acta Crystallogr.,Sect.D, 65, 2009
|
|
7B1X
 
 | | Crystal structure of cold-active esterase PMGL3 from permafrost metagenomic library | | 分子名称: | esterase PMGL3 | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Petrovskaya, L.E, Kryukova, M.V, Kryukova, E.A, Korzhenevsky, D.A, Lomakina, G.Y, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | 登録日 | 2020-11-25 | | 公開日 | 2021-11-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Biochemical Characterization of a Cold-Active PMGL3 Esterase with Unusual Oligomeric Structure.
Biomolecules, 11, 2021
|
|
7ARZ
 
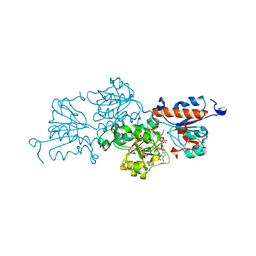 | | Ternary complex of NAD-dependent formate dehydrogenase from Physcomitrium patens | | 分子名称: | AZIDE ION, Formate dehydrogenase, mitochondrial, ... | | 著者 | Goryaynova, D.A, Nikolaeva, A.Y, Pometun, A.A, Savin, S.S, Parshin, P.D, Popov, V.O, Tishkov, V.I, Boyko, K.M. | | 登録日 | 2020-10-26 | | 公開日 | 2021-11-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Ternary complex of NAD-dependent formate dehydrogenase from Physcomitrium patens
To Be Published
|
|
3UU9
 
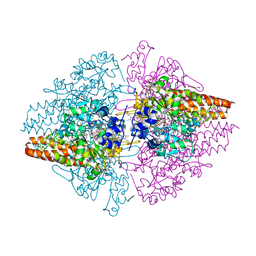 | | Structure of the free TvNiRb form of Thioalkalivibrio nitratireducens cytochrome c nitrite reductase | | 分子名称: | CALCIUM ION, Eight-heme nitrite reductase, HEME C, ... | | 著者 | Trofimov, A.A, Polyakov, K.M, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | 登録日 | 2011-11-28 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8RAF
 
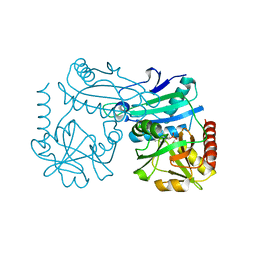 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis point mutant R90I (holo form) | | 分子名称: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y, Boyko, K.M. | | 登録日 | 2023-12-01 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Multifunctionality of arginine residues in the active sites of non-canonical d-amino acid transaminases.
Arch.Biochem.Biophys., 756, 2024
|
|
8RAI
 
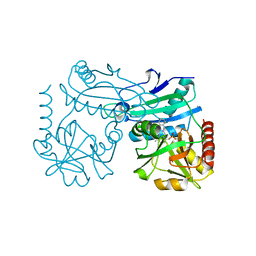 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis point mutant R90I complexed with phenylhydrazine | | 分子名称: | Aminotransferase class IV, GLYCEROL, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | 著者 | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y, Boyko, K.M. | | 登録日 | 2023-12-01 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Multifunctionality of arginine residues in the active sites of non-canonical d-amino acid transaminases.
Arch.Biochem.Biophys., 756, 2024
|
|
3TN7
 
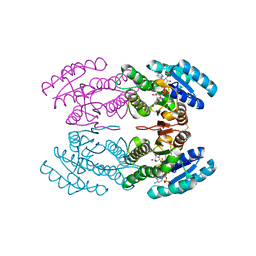 | | Crystal structure of short-chain alcohol dehydrogenase from hyperthermophilic archaeon Thermococcus sibiricus complexed with 5-hydroxy-NADP | | 分子名称: | 5-hydroxy-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, GLYCEROL, Short-chain alcohol dehydrogenase | | 著者 | Boyko, K.M, Polyakov, K.M, Bezsudnova, E.Y, Stekhanova, T.N, Gumerov, V.M, Mardanov, A.V, Ravin, N.V, Skryabin, K.G, Kovalchuk, M.V, Popov, V.O. | | 登録日 | 2011-09-01 | | 公開日 | 2012-08-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structural insight into the molecular basis of polyextremophilicity of short-chain alcohol dehydrogenase from the hyperthermophilic archaeon Thermococcus sibiricus.
Biochimie, 94, 2012
|
|
4YJK
 
 | | Crystal structure of C212S mutant of Shewanella oneidensis MR-1 uridine phosphorylase | | 分子名称: | SULFATE ION, URACIL, Uridine phosphorylase | | 著者 | Safonova, T.N, Mordkovich, N.N, Manuvera, V.A, Dorovatovsky, P.V, Veiko, V.P, Popov, V.O, Polyakov, K.M. | | 登録日 | 2015-03-03 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Concerted action of two subunits of the functional dimer of Shewanella oneidensis MR-1 uridine phosphorylase derived from a comparison of the C212S mutant and the wild-type enzyme.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6TTB
 
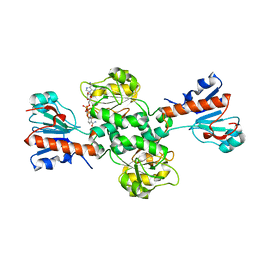 | | Crystal structure of NAD-dependent formate dehydrogenase from Staphylococcus aureus in complex with NAD | | 分子名称: | Formate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Boyko, K.M, Pometun, A.A, Nikolaeva, A.Y, Kargov, I.S, Yurchenko, T.S, Savin, S.S, Popov, V.O, Tishkov, V.I. | | 登録日 | 2019-12-26 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of NAD-dependent formate dehydrogenase from Staphylococcus aureus in complex with NAD
To Be Published
|
|
3RKH
 
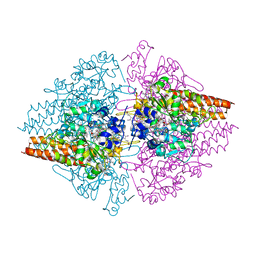 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with nitrite (full occupancy) | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, Eight-heme nitrite reductase, ... | | 著者 | Trofimov, A.A, Polyakov, K.M, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | 登録日 | 2011-04-18 | | 公開日 | 2011-06-15 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RR5
 
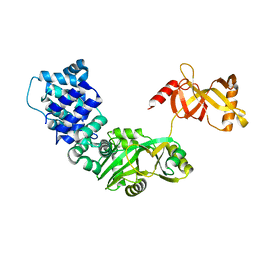 | | DNA ligase from the archaeon Thermococcus sp. 1519 | | 分子名称: | DNA ligase, MAGNESIUM ION | | 著者 | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Popov, V.O, Polyakov, K.M, Ravin, N.V, Shabalin, I.G, Skryabin, K.G, Stekhanova, T.N, Kovalchuk, M.V. | | 登録日 | 2011-04-29 | | 公開日 | 2012-04-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.018 Å) | | 主引用文献 | ATP-dependent DNA ligase from Thermococcus sp. 1519 displays a new arrangement of the OB-fold domain.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3OWM
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with hydroxylamine | | 分子名称: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | 著者 | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Tikhonova, T.V, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | 登録日 | 2010-09-20 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Binding of sulfite by the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase
To be Published
|
|
3S7W
 
 | | Structure of the TvNiRb form of Thioalkalivibrio nitratireducens cytochrome c nitrite reductase with an oxidized Gln360 in a complex with hydroxylamine | | 分子名称: | AZIDE ION, CALCIUM ION, Eight-heme nitrite reductase, ... | | 著者 | Trofimov, A.A, Polyakov, K.M, Tikhonova, T.V, Popov, V.O. | | 登録日 | 2011-05-27 | | 公開日 | 2011-07-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structure of the TvNiRb form of Thioalkalivibrio nitratireducens cytochrome c nitrite reductase with an oxidized Gln360 in a complex with hydroxylamine
To be Published
|
|
3SCE
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase with a covalent bond between the CE1 atom of Tyr303 and the CG atom of Gln360 (TvNiRb) | | 分子名称: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | 著者 | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | 登録日 | 2011-06-07 | | 公開日 | 2011-07-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8Z77
 
 | | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum (pmTcDH), activated by crystals soaking with 1 mM CuCl2 and Na ascorbate during 12 hours | | 分子名称: | 1,2-ETHANEDIOL, COPPER (II) ION, Twin-arginine translocation signal domain-containing protein | | 著者 | Varfolomeeva, L.A, Solovieva, A.Y, Shipkov, N.S, Dergousova, N.I, Minyaev, M.E, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | 登録日 | 2024-04-19 | | 公開日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum (pmTcDH), activated by crystals soaking with 1 mM CuCl2 and Na ascorbate during 12 hours
To Be Published
|
|
8Z75
 
 | | The structure of non-activated thiocyanate dehydrogenase from Pelomicrobium methylotrophicum (pmTcDH) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | 著者 | Varfolomeeva, L.A, Solovieva, A.Y, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | 登録日 | 2024-04-19 | | 公開日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The structure of non-activated thiocyanate dehydrogenase from Pelomicrobium methylotrophicum (pmTcDH)
To Be Published
|
|
8Z76
 
 | | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum (pmTcDH), activated by crystals soaking with 1 mM CuCl2 during 6 months | | 分子名称: | 1,2-ETHANEDIOL, COPPER (II) ION, SODIUM ION, ... | | 著者 | Varfolomeeva, L.A, Solovieva, A.Y, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | 登録日 | 2024-04-19 | | 公開日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum (pmTcDH), activated by crystals soaking with 1 mM CuCl2
To Be Published
|
|
8ZES
 
 | | Crystal structure of the Wuhan SARS-CoV-2 RBD (333-541) complexed with P2C5 nanobody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody P2C5, ... | | 著者 | Sluchanko, N.N, Varfolomeeva, L.A, Shcheblyakov, D.V, Logunov, D.Y, Gintsburg, A.L, Popov, V.O, Boyko, K.M. | | 登録日 | 2024-05-06 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structural Basis for Evasion of New SARS-CoV-2 Variants from the Potent Virus-Neutralizing Nanobody Targeting the S-Protein Receptor-Binding Domain.
Biochemistry Mosc., 89, 2024
|
|
8ZER
 
 | | Crystal structure of the complex of Wuhan SARS-CoV-2 RBD (319-541) with P2C5 nanobody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody P2C5, Spike protein S1, ... | | 著者 | Sluchanko, N.N, Varfolomeeva, L.A, Shcheblyakov, D.V, Logunov, D.Y, Gintsburg, A.L, Popov, V.O, Boyko, K.M. | | 登録日 | 2024-05-06 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Basis for Evasion of New SARS-CoV-2 Variants from the Potent Virus-Neutralizing Nanobody Targeting the S-Protein Receptor-Binding Domain.
Biochemistry Mosc., 89, 2024
|
|
5N1T
 
 | | Crystal structure of complex between flavocytochrome c and copper chaperone CopC from T. paradoxus | | 分子名称: | COPPER (II) ION, CopC, Cytochrome C, ... | | 著者 | Osipov, E.M, Lilina, A.V, Tikhonova, T.V, Tsallagov, S.I, Popov, V.O. | | 登録日 | 2017-02-06 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of the flavocytochrome c sulfide dehydrogenase associated with the copper-binding protein CopC from the haloalkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio paradoxusARh 1.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5MWC
 
 | | Crystal structure of the genetically-encoded green calcium indicator NTnC in its calcium bound state | | 分子名称: | CALCIUM ION, genetically-encoded green calcium indicator NTnC | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Rakitina, T.V, Popov, V.O, Subach, O.M, Barykina, N.V, Subach, F.V. | | 登録日 | 2017-01-18 | | 公開日 | 2018-02-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Enchanced variant of genetically-encoded green calcium indicator NTnC
To Be Published
|
|
5OEX
 
 | | Complex with iodine ion for thiocyanate dehydrogenase from Thioalkalivibrio paradoxus | | 分子名称: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, COPPER (II) ION, ... | | 著者 | Polyakov, K.M, Tsallagov, S.I, Tikhonova, T.V, Popov, V.O. | | 登録日 | 2017-07-10 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery and characterization of a novel copper containing enzyme - THIOCYANATE DEHYDROGENASE.
To Be Published
|
|
6ET6
 
 | | Crystal structure of muramidase from Acinetobacter baumannii AB 5075UW prophage | | 分子名称: | GLYCEROL, Lysozyme, SULFATE ION | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Sykilinda, N.N, Shneider, M.M, Miroshnikov, K.A, Popov, V.O. | | 登録日 | 2017-10-25 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure of anAcinetobacterBroad-Range Prophage Endolysin Reveals a C-Terminal alpha-Helix with the Proposed Role in Activity against Live Bacterial Cells.
Viruses, 10, 2018
|
|
