4JGN
 
 | |
4JNX
 
 | |
4J5V
 
 | |
4JK0
 
 | |
4KNQ
 
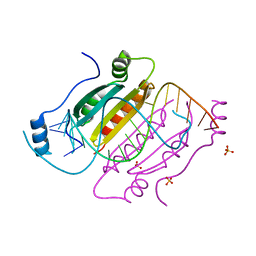 | |
2HLN
 
 | | L-asparaginase from Erwinia carotovora in complex with glutamic acid | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTAMIC ACID, L-asparaginase | | Authors: | Kravchenko, O.V, Kislitsin, Y.A, Popov, A.N, Nikonov, S.V, Kuranova, I.P. | | Deposit date: | 2006-07-08 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of L-asparaginase from Erwinia carotovora complexed with aspartate and glutamate.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4GHS
 
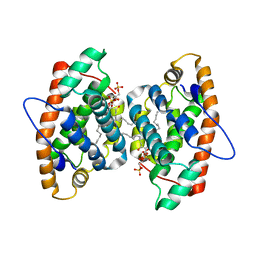 | | Crystal structure of human GLTP bound with 12:0 disulfatide (orthorombic form; two subunits in asymmetric unit) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Popov, A.N, Goni-de-Cerio, F, Cabo-Bilbao, A, Malinina, L. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GHP
 
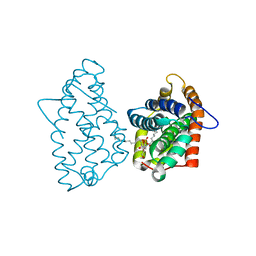 | | Crystal Structure of D48V||A47D mutant of Human GLTP bound with 12:0 monosulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-08 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8OUC
 
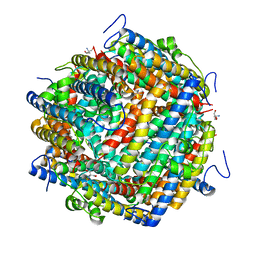 | | Escherichia coli DPS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA protection during starvation protein | | Authors: | Kovalenko, V.V, Loiko, N.G, Tereshkina, K.B, Tereshkin, E.V, Chulichkov, A.L, Popov, A.N, Krupyanskii, Y.F. | | Deposit date: | 2023-04-22 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Escherichia coli DPS
To be published
|
|
4GXG
 
 | | Crystal structure of human GLTP bound with 12:0 monosulfatide (orthorhombic form; four subunits in asymmetric unit) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Goni-de-Cerio, F, Popov, A.N, Malinina, L. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GVT
 
 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 disulfatide (hexagonal form) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Goni-de-Cerio, F, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-31 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2GVN
 
 | | L-asparaginase from Erwinia carotovora in complex with aspartic acid | | Descriptor: | ASPARTIC ACID, L-asparaginase | | Authors: | Kravchenko, O.V, Kislitsin, Y.A, Popov, A.N, Nikonov, S.V, Kuranova, I.P. | | Deposit date: | 2006-05-03 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-dimensional structures of L-asparaginase from Erwinia carotovora complexed with aspartate and glutamate.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4GJQ
 
 | | Crystal structure of human GLTP bound with 12:0 monosulfatide (orthorhombic form;two subunits in asymmetric unit) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein, HEXANE | | Authors: | Cabo-Bilbao, A, Goni-de-Cerio, F, Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-10 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GH0
 
 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 monosulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide, PENTADECANE | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-07 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GXD
 
 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 disulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2BI1
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure B) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BI5
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure E) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2CNU
 
 | | Atomic resolution structure of SAICAR-synthase from Saccharomyces cerevisiae complexed with aspartic acid | | Descriptor: | ASPARTIC ACID, PHOSPHORIBOSYLAMINOIMIDAZOLE-SUCCINOCARBOXAMIDE SYNTHASE, SULFATE ION | | Authors: | Urusova, D.V, Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Barynin, V.V, Popov, A.N, Lamzin, V.S, Melik-Adamyan, W.R. | | Deposit date: | 2006-05-24 | | Release date: | 2006-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Saicar Synthase: Substrate Recognition, Conformational Flexibility and Catalysis.
To be Published
|
|
2CNQ
 
 | | Atomic resolution structure of SAICAR-synthase from Saccharomyces cerevisiae complexed with ADP, AICAR, succinate | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | Authors: | Urusova, D.V, Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Barynin, V.V, Popov, A.N, Lamzin, V.S, Melik-Adamyan, W.R. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Saicar Synthase: Substrate Recognition, Conformational Flexibility and Catalysis.
To be Published
|
|
2BI2
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure C) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BIE
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure H) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-21 | | Release date: | 2005-05-19 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BI9
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure F) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BIA
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure G) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BHX
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure A) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BIG
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure I) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-21 | | Release date: | 2005-05-19 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
