5A76
 
 | |
2J98
 
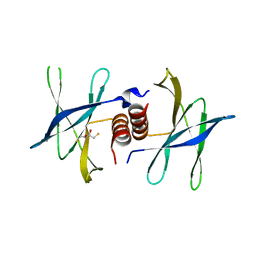 | | Human coronavirus 229E non structural protein 9 cys69ala mutant (Nsp9) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, REPLICASE POLYPROTEIN 1AB | | Authors: | Ponnusamy, R, Mesters, J.R, Moll, R, Hilgenfeld, R. | | Deposit date: | 2006-11-03 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Variable Oligomerization Modes in Coronavirus Non-Structural Protein 9.
J.Mol.Biol., 383, 2008
|
|
2J97
 
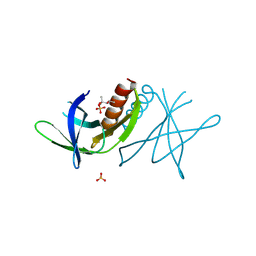 | | Human coronavirus 229E non structural protein 9 (Nsp9) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, REPLICASE POLYPROTEIN 1AB, SULFATE ION | | Authors: | Ponnusamy, R, Mesters, J.R, Moll, R, Hilgenfeld, R. | | Deposit date: | 2006-11-03 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Variable Oligomerization Modes in Coronavirus Non-Structural Protein 9.
J.Mol.Biol., 383, 2008
|
|
4B92
 
 | |
4CNU
 
 | |
4B91
 
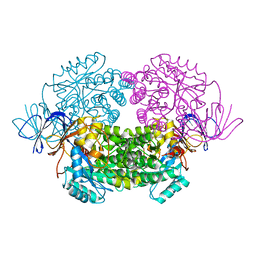 | | Crystal structure of truncated human CRMP-5 | | Descriptor: | DIHYDROPYRIMIDINASE-RELATED PROTEIN 5 | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2012-08-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights Into the Oligomerization of Crmps: Crystal Structure of Human Collapsin Response Mediator Protein 5.
J.Neurochem., 125, 2013
|
|
4CNS
 
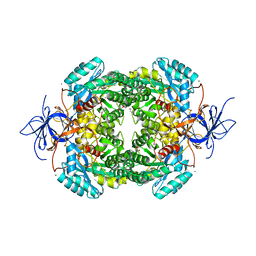 | | Crystal structure of truncated human CRMP-4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2014-01-24 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Crmp-4: Correction of Intensities for Lattice-Translocation Disorder
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CNT
 
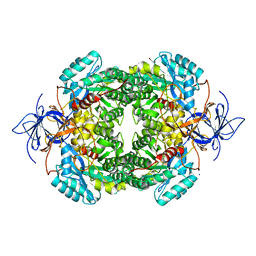 | | CRYSTAL STRUCTURE OF WT HUMAN CRMP-4 | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROPYRIMIDINASE-LIKE 3, SODIUM ION | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2014-01-24 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of Human Crmp-4: Correction of Intensities for Lattice-Translocation Disorder
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4B90
 
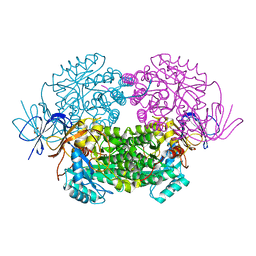 | | Crystal structure of WT human CRMP-5 | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROPYRIMIDINASE-RELATED PROTEIN 5 | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2012-08-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights Into the Oligomerization of Crmps: Crystal Structure of Human Collapsin Response Mediator Protein 5.
J.Neurochem., 125, 2013
|
|
3O5Z
 
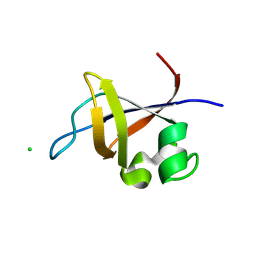 | | Crystal structure of the SH3 domain from p85beta subunit of phosphoinositide 3-kinase (PI3K) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Chen, S, Xiao, Y, Ponnusamy, R, Tan, J, Lei, J, Hilgenfeld, R. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | X-ray structure of the SH3 domain of the phosphoinositide 3-kinase p85 beta subunit
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3P31
 
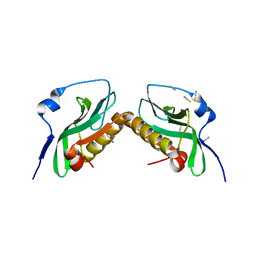 | |
4BLG
 
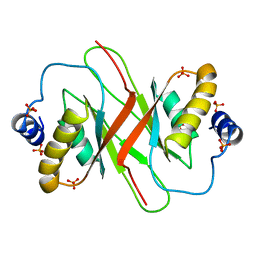 | | Crystal structure of MHV-68 Latency-associated nuclear antigen (LANA) C-terminal DNA binding domain | | Descriptor: | LATENCY-ASSOCIATED NUCLEAR ANTIGEN, PHOSPHATE ION | | Authors: | Correia, B, Cerqueira, S.A, Beauchemin, C, Pires De Miranda, M, Li, S, Ponnusamy, R, Rodrigues, L, Schneider, T.R, Carrondo, M.A, Kaye, K.M, Simas, J.P, McVey, C.E. | | Deposit date: | 2013-05-02 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Gamma-2 Herpesvirus Lana DNA Binding Domain Identifies Charged Surface Residues which Impact Viral Latency
Plos Pathog., 9, 2013
|
|
