6RHH
 
 | | Single crystal serial study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi at sub-atomic resolution. Third structure of the series with 315 KGy dose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Laccase 2, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2019-04-20 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The subatomic resolution study of laccase inhibition by chloride and fluoride anions using single-crystal serial crystallography: insights into the enzymatic reaction mechanism.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6RHX
 
 | | Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by chloride anions at sub-atomic resolution. Third structure of the series with 315 KGy dose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2019-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | The subatomic resolution study of laccase inhibition by chloride and fluoride anions using single-crystal serial crystallography: insights into the enzymatic reaction mechanism.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6RI6
 
 | | Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. Second structure of the series with 400 KGy dose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, FLUORIDE ION, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2019-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | The subatomic resolution study of laccase inhibition by chloride and fluoride anions using single-crystal serial crystallography: insights into the enzymatic reaction mechanism.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6RIL
 
 | | Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. Fourteenth structure of the series with 5600 KGy dose (data was collected after refreezing). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, FLUORIDE ION, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2019-04-24 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The subatomic resolution study of laccase inhibition by chloride and fluoride anions using single-crystal serial crystallography: insights into the enzymatic reaction mechanism.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6RGH
 
 | | Single crystal serial study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi at sub-atomic resolution. First structure of the series with 15 KGy dose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Laccase 2, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2019-04-16 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The subatomic resolution study of laccase inhibition by chloride and fluoride anions using single-crystal serial crystallography: insights into the enzymatic reaction mechanism.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6RHI
 
 | | Single crystal serial study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi at sub-atomic resolution. Ninth structure of the series with 1215 KGy dose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Laccase 2, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2019-04-20 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | The subatomic resolution study of laccase inhibition by chloride and fluoride anions using single-crystal serial crystallography: insights into the enzymatic reaction mechanism.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6G50
 
 | | The structure of thiocyanate dehydrogenase from Thioalkalivibrio paradoxus as isolated. | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, SULFATE ION, ... | | Authors: | Polyakov, K.M, Tsallagov, S.I, Tikhkonova, T.V, Popov, V.O. | | Deposit date: | 2018-03-28 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4F8X
 
 | | Penicillium canescens endo-1,4-beta-xylanase XylE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Endo-1,4-beta-xylanase, beta-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Polyakov, K.M, Trofimov, A.A, Fedorova, T.V, Koroleva, O.V, Maisuradze, I.G, Chulkin, A.M, Vavilova, E.A, Benevolenskii, S.V. | | Deposit date: | 2012-05-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: |
|
|
6SJI
 
 | | The structure of thiocyanate dehydrogenase from Thioalkalivibrio paradoxus mutant with His 482 replaced by Gln | | Descriptor: | COPPER (II) ION, SULFATE ION, thiocyanate dehydrogenase | | Authors: | Polyakov, K.M, Tikhonova, T.V, Rakitina, T.V, Osipov, E, Popov, V.O. | | Deposit date: | 2019-08-13 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8YOU
 
 | | The pmTcDH complex structure with an inhibitor SeCN | | Descriptor: | COPPER (II) ION, GLYCEROL, SELENIUM ATOM, ... | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-13 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The pmTcDH complex structure with an inhibitor SeCN
To Be Published
|
|
3GM6
 
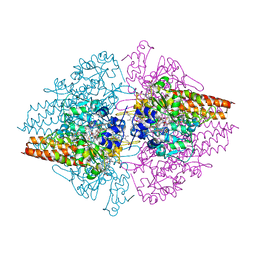 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in complex with phosphate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Filimonenkov, A.A, Dorovatovsky, P.V, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2009-03-13 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of octaheme cytochrome c nitrite reductase from Thioalkalivibrio nitratireducens in a complex with phosphate
Crystallography Reports, 55, 2010
|
|
3LGQ
 
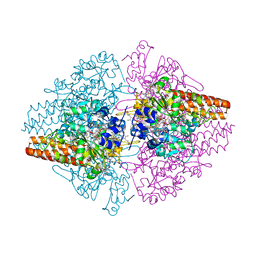 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in complex with sulfite (modified Tyr-303) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Filimonenkov, A.A, Tikhonova, T.V, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2010-01-21 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3LG1
 
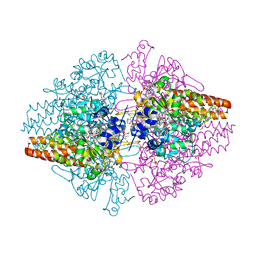 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase reduced by sodium borohydride (in complex with sulfite) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Filimonenkov, A.A, Dorovatovsky, P.V, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2010-01-19 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3MMO
 
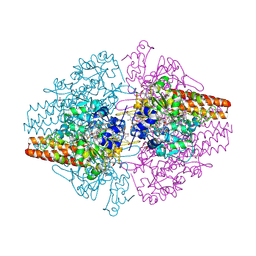 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in complex with cyanide | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Tikhonova, T.V, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2010-04-20 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of complexes of octahaem cytochrome c nitrite reductase from Thioalkalivibrio nitratireducens with sulfite and cyanide.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5EK6
 
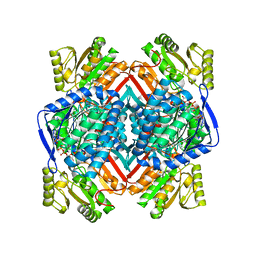 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 complexed with NADP and isobutyraldehyde | | Descriptor: | 2-methylpropanal, Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Polyakov, K.M, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
2GUG
 
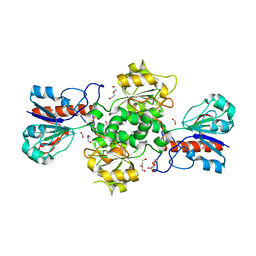 | | NAD-dependent formate dehydrogenase from Pseudomonas sp.101 in complex with formate | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Formate dehydrogenase, ... | | Authors: | Filippova, E.V, Polyakov, K.M, Tikhonova, T.V, Boiko, K.M, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2006-04-30 | | Release date: | 2006-05-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the complex of NAD-dependent formate dehydrogenase from
metylotrophic bacterium Pseudomonas sp.101 with formate.
KRISTALLOGRAFIYA, 51, 2006
|
|
2GO1
 
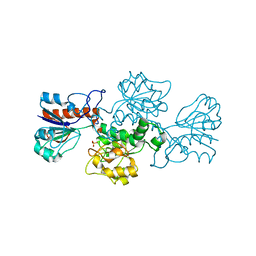 | | NAD-dependent formate dehydrogenase from Pseudomonas sp.101 | | Descriptor: | NAD-dependent formate dehydrogenase, SULFATE ION | | Authors: | Filippova, E.V, Polyakov, K.M, Tikhonova, T.V, Stekhanova, T.N, Boiko, K.M, Popov, V.O. | | Deposit date: | 2006-04-12 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a new crystal modification of the bacterial NAD-dependent formate dehydrogenase with a resolution of 2.1 A
Crystallography reports, 50, 2005
|
|
8P3L
 
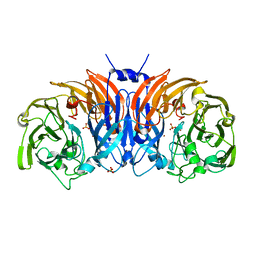 | | The structure of thiocyanate dehydrogenase mutant form with Thr 169 replaced by Ala from Thioalkalivibrio paradoxus | | Descriptor: | COPPER (II) ION, SULFATE ION, Twin-arginine translocation signal domain-containing protein | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Komolov, A.S, Rakitina, T.V, Dergousova, N.I, Dorovatovskii, P.V, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-05-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Improvement of the Diffraction Properties of Thiocyanate Dehydrogenase Crystals
Crystallography Reports, 2023
|
|
8P3M
 
 | | The structure of thiocyanate dehydrogenase mutant form with Lys 281 replaced by Ala from Thioalkalivibrio paradoxus | | Descriptor: | BORIC ACID, COPPER (II) ION, SODIUM ION, ... | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Komolov, A.S, Rakitina, T.V, Dergousova, N.I, Dorovatovskii, P.V, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-05-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Improvement of the Diffraction Properties of Thiocyanate Dehydrogenase Crystals
Crystallography Reports, 2023
|
|
8Q9X
 
 | | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum with molecular oxygen at 1.05 A resolution | | Descriptor: | COPPER (II) ION, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-08-22 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum at atomic resolution
To Be Published
|
|
8Q9Y
 
 | | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum in complex with inhibitor thiourea at 1.10 A resolution | | Descriptor: | COPPER (II) ION, GLYCEROL, THIOUREA, ... | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-08-22 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum at atomic resolution
To Be Published
|
|
3N7U
 
 | | NAD-dependent formate dehydrogenase from higher-plant Arabidopsis thaliana in complex with NAD and azide | | Descriptor: | AZIDE ION, Formate dehydrogenase, GLYCEROL, ... | | Authors: | Shabalin, I.G, Polyakov, K.M, Serov, A.E, Skirgello, O.E, Sadykhov, E.G, Dorovatovskiy, P.V, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the apo and holo forms of NAD-dependent formate dehydrogenase from the higher-plant Arabidopsis Thaliana
To be Published
|
|
1XEY
 
 | | Crystal structure of the complex of Escherichia coli GADA with glutarate at 2.05 A resolution | | Descriptor: | ACETATE ION, GLUTARIC ACID, Glutamate decarboxylase alpha, ... | | Authors: | Dutyshev, D.I, Darii, E.L, Fomenkova, N.P, Pechik, I.V, Polyakov, K.M, Nikonov, S.V, Andreeva, N.S, Sukhareva, B.S. | | Deposit date: | 2004-09-13 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Escherichia coli glutamate decarboxylase (GADalpha) in complex with glutarate at 2.05 angstroms resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3NAQ
 
 | | Apo-form of NAD-dependent formate dehydrogenase from higher-plant Arabidopsis thaliana | | Descriptor: | Formate dehydrogenase, SULFATE ION | | Authors: | Shabalin, I.G, Polyakov, K.M, Serov, A.E, Skirgello, O.E, Sadykhov, E.G, Dorovatovskiy, P.V, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the apo and holo forms of NAD-dependent formate dehydrogenase from the higher-plant Arabidopsis Thaliana
to be published
|
|
3JTM
 
 | | Structure of recombinant formate dehydrogenase from Arabidopsis thaliana | | Descriptor: | AZIDE ION, Formate dehydrogenase, mitochondrial, ... | | Authors: | Timofeev, V.I, Shabalin, I.G, Serov, A.E, Polyakov, K.M, Popov, V.O, Tishkov, V.I, Kuranova, I.P, Samigina, V.R. | | Deposit date: | 2009-09-13 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of recombinant formate dehydrogenase from Arabidopsis thaliana
to be published
|
|
