6EI1
 
 | | Crystal structure of the covalent complex between deubiquitinase ZUFSP (ZUP1) and Ubiquitin-PA | | Descriptor: | GLYCEROL, MALONATE ION, Polyubiquitin-B, ... | | Authors: | Pichlo, C, Baumann, U, Hofmann, K, Hermanns, T. | | Deposit date: | 2017-09-16 | | Release date: | 2018-03-07 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | A family of unconventional deubiquitinases with modular chain specificity determinants.
Nat Commun, 9, 2018
|
|
7OIY
 
 | | Crystal structure of the ZUFSP family member Mug105 | | Descriptor: | SODIUM ION, Ubiquitin carboxyl-terminal hydrolase mug105 | | Authors: | Pichlo, C, Hermanns, T, Hofmann, K, Baumann, U. | | Deposit date: | 2021-05-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structural basis for the diverse linkage specificities within the ZUFSP deubiquitinase family.
Nat Commun, 13, 2022
|
|
7NZY
 
 | | Crystal structure of human Casein Kinase I delta in complex with CGS-15943 | | Descriptor: | 9-chloranyl-2-(furan-2-yl)-[1,2,4]triazolo[1,5-c]quinazolin-5-amine, Casein kinase I isoform delta, MALONATE ION, ... | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2021-03-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Phenotypic Discovery of Triazolo[1,5- c ]quinazolines as a First-In-Class Bone Morphogenetic Protein Amplifier Chemotype.
J.Med.Chem., 65, 2022
|
|
7OJE
 
 | | Crystal structure of the covalent complex between Tribolium castaneum deubiquitinase ZUP and Ubiquitin-PA | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Lys-63-specific deubiquitinase ZUFSP, ... | | Authors: | Pichlo, C, Hermanns, T, Hofmann, K, Baumann, U. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structural basis for the diverse linkage specificities within the ZUFSP deubiquitinase family.
Nat Commun, 13, 2022
|
|
6FSM
 
 | | Crystal structure of TCE-treated Thermolysin | | Descriptor: | CALCIUM ION, GLYCEROL, LYSINE, ... | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2018-02-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Improved protein-crystal identification by using 2,2,2-trichloroethanol as a fluorescence enhancer.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6HMP
 
 | | Crystal structure of human Casein Kinase I delta in complex with a photoswitchable 2-Azoimidazole-based Inhibitor (compound 3) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[5-(4-fluorophenyl)-2-[(~{E})-(4-fluorophenyl)diazenyl]-3-methyl-imidazol-4-yl]pyridin-2-yl]propanamide, Casein kinase I isoform delta, PHOSPHATE ION | | Authors: | Pichlo, C, Schehr, M, Charl, J, Brunstein, E, Peifer, C, Baumann, U. | | Deposit date: | 2018-09-12 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
6HMR
 
 | | Crystal structure of human Casein Kinase I delta in complex with a photoswitchable 2-Azothiazole-based inhibitor (compound 2) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-[(~{E})-phenyldiazenyl]-1,3-thiazol-5-yl]pyridin-2-yl]propanamide, Casein kinase I isoform delta, MALONIC ACID | | Authors: | Pichlo, C, Schehr, M, Charl, J, Brunstein, E, Peifer, C, Baumann, U. | | Deposit date: | 2018-09-12 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
6F26
 
 | | Crystal structure of human Casein Kinase I delta in complex with compound 31b | | Descriptor: | (9~{S},10~{S},11~{R})-~{N}-[4-[3-(4-fluorophenyl)-5-propan-2-yl-1,2-oxazol-4-yl]pyridin-2-yl]-4-(4-methoxyphenyl)-10,11-bis(oxidanyl)-1,7-diazatricyclo[7.3.0.0^{3,7}]dodeca-3,5-diene-6-carboxamide, Casein kinase I isoform delta, SULFATE ION | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2017-11-23 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design, Synthesis and Biological Evaluation of Isoxazole-Based CK1 Inhibitors Modified with Chiral Pyrrolidine Scaffolds.
Molecules, 24, 2019
|
|
5N12
 
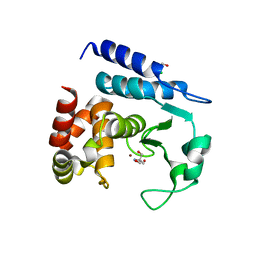 | | Crystal structure of TCE treated rPPEP-1 | | Descriptor: | 2,2,2-tris-chloroethanol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ... | | Authors: | Pichlo, C, Schacherl, M, Baumann, U. | | Deposit date: | 2017-02-04 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Improved protein-crystal identification by using 2,2,2-trichloroethanol as a fluorescence enhancer.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6FSJ
 
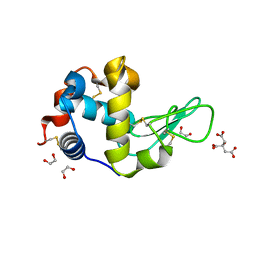 | | Crystal structure of TCE-treated Lysozyme | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Lysozyme C | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2018-02-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Improved protein-crystal identification by using 2,2,2-trichloroethanol as a fluorescence enhancer.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6F1W
 
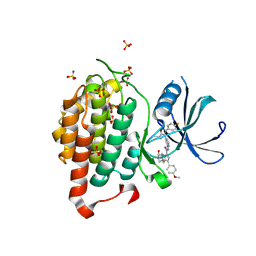 | | Crystal structure of human Casein Kinase I delta in complex with compound 31a | | Descriptor: | (9~{R},10~{R},11~{S})-~{N}-[4-[3-(4-fluorophenyl)-5-propan-2-yl-1,2-oxazol-4-yl]pyridin-2-yl]-4-(4-methoxyphenyl)-10,11-bis(oxidanyl)-1,7-diazatricyclo[7.3.0.0^{3,7}]dodeca-3,5-diene-6-carboxamide, Casein kinase I isoform delta, SULFATE ION | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2017-11-23 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | Design, Synthesis and Biological Evaluation of Isoxazole-Based CK1 Inhibitors Modified with Chiral Pyrrolidine Scaffolds.
Molecules, 24, 2019
|
|
5OKT
 
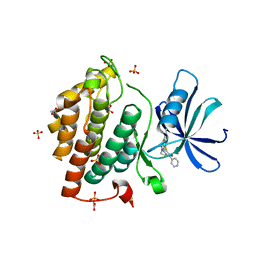 | | Crystal structure of human Casein Kinase I delta in complex with IWP-2 | | Descriptor: | ACETATE ION, Casein kinase I isoform delta, GLYCEROL, ... | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of Inhibitor of Wnt Production 2 (IWP-2) and Related Compounds As Selective ATP-Competitive Inhibitors of Casein Kinase 1 (CK1) delta / epsilon.
J. Med. Chem., 61, 2018
|
|
5MQV
 
 | | Crystal structure of human Casein Kinase I delta in complex with 4-(2,5-Dimethoxyphenyl)-N-(4-(5-(4-fluorphenyl)-2-(methylthio)-1H-imidazol-4-yl)-pyridin-2-yl)-1-methyl-1H-pyrrole-2-carboxamide | | Descriptor: | 4-(2,5-Dimethoxyphenyl)-N-(4-(5-(4-fluorphenyl)-2-(methylthio)-1H-imidazol-4-yl)-pyridin-2-yl)-1-methyl-1H-pyrrole-2-carboxamide, Casein kinase I isoform delta, PHOSPHATE ION | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2016-12-20 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Optimized 4,5-Diarylimidazoles as Potent/Selective Inhibitors of Protein Kinase CK1 delta and Their Structural Relation to p38 alpha MAPK.
Molecules, 22, 2017
|
|
6R56
 
 | | Crystal structure of PPEP-1(K101E/E184K) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ZINC ION | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2019-03-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Molecular determinants of the mechanism and substrate specificity ofClostridium difficileproline-proline endopeptidase-1.
J.Biol.Chem., 294, 2019
|
|
6R4Z
 
 | |
6R54
 
 | | Crystal structure of PPEP-1(E184A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NICKEL (II) ION, Pro-Pro endopeptidase, ... | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2019-03-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.417 Å) | | Cite: | Molecular determinants of the mechanism and substrate specificity ofClostridium difficileproline-proline endopeptidase-1.
J.Biol.Chem., 294, 2019
|
|
6R5A
 
 | | Crystal structure of PPEP-1(W103F) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ZINC ION | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2019-03-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Molecular determinants of the mechanism and substrate specificity ofClostridium difficileproline-proline endopeptidase-1.
J.Biol.Chem., 294, 2019
|
|
6R55
 
 | | Crystal structure of PPEP-1(E184K) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ZINC ION | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2019-03-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Molecular determinants of the mechanism and substrate specificity ofClostridium difficileproline-proline endopeptidase-1.
J.Biol.Chem., 294, 2019
|
|
6R52
 
 | | Crystal structure of PPEP-1(K101A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ZINC ION | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2019-03-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | Molecular determinants of the mechanism and substrate specificity ofClostridium difficileproline-proline endopeptidase-1.
J.Biol.Chem., 294, 2019
|
|
6R53
 
 | | Crystal structure of PPEP-1(K101R) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ZINC ION | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2019-03-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Molecular determinants of the mechanism and substrate specificity ofClostridium difficileproline-proline endopeptidase-1.
J.Biol.Chem., 294, 2019
|
|
6R4Y
 
 | |
6R5B
 
 | |
6R51
 
 | |
6R58
 
 | |
6R57
 
 | |
