6UBL
 
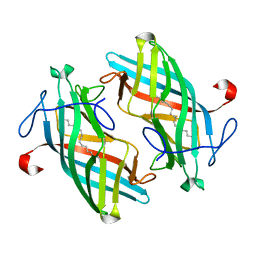 | | Structure of DynF from the Dynemicin Biosynthesis Pathway of Micromonospora chersina | | Descriptor: | DynF, PALMITIC ACID | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Bhardwaj, M, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
3DHI
 
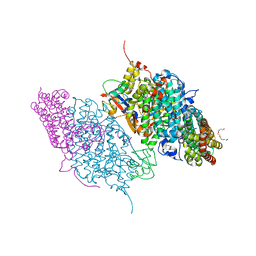 | | Crystal Structure of Reduced Toluene 4-Monoxygenase Hydroxylase Complexed with Effector Protein | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, FE (III) ION, ... | | Authors: | Bailey, L.J, Mccoy, J.G, Phillips Jr, G.N, Fox, B.G. | | Deposit date: | 2008-06-17 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural consequences of effector protein complex formation in a diiron hydroxylase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6UVQ
 
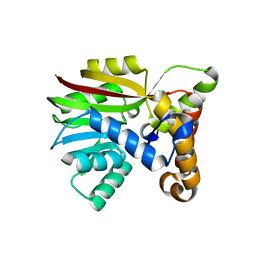 | | Crystal structure of Apo AtmM | | Descriptor: | ACETATE ION, D-glucose O-methyltransferase, MAGNESIUM ION | | Authors: | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-11-04 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of Apo AtmM
To Be Published
|
|
6UWD
 
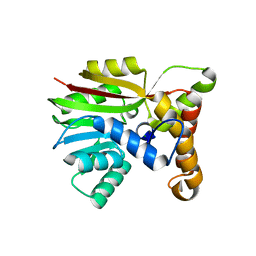 | | Crystal structure of Apo AtmM | | Descriptor: | ACETATE ION, D-glucose O-methyltransferase, MAGNESIUM ION | | Authors: | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-11-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of Apo AtmM
To Be Published
|
|
6UPP
 
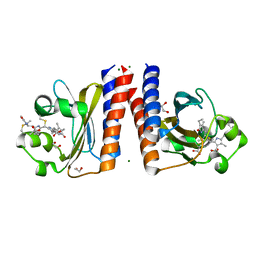 | | Radiation Damage Test of PixJ Pb state crystals | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Clinger, J.A, Miller, M.D, Burgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UV6
 
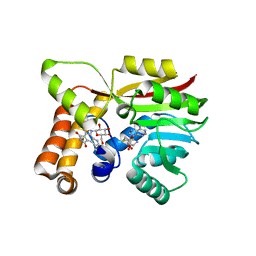 | | AtmM with bound rebeccamycin analogue | | Descriptor: | 12-beta-D-glucopyranosyl-12,13-dihydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazole-5,7(6H)-dione, D-glucose O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of AtmM Bound with Glycosylated Indolocarbazole
To Be Published
|
|
6V04
 
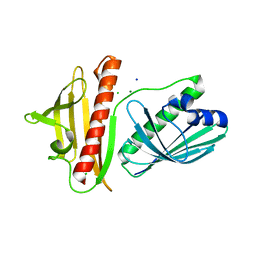 | | DynU16 crystal structure, a putative protein in the dynemicin biosynthetic locus | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Alvarado, S.K, Miller, M.D, Bhardwaj, M, Thorson, J.S, Van Lanen, S.G, Phillips Jr, G.N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of DynU16, a START/Bet v1-like protein involved in dynemicin biosynthesis.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6VQP
 
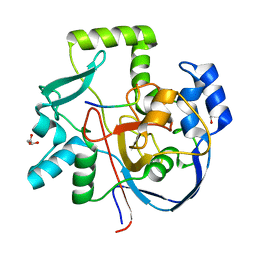 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CalU17, CalU17 His-Tagged protein, GLYCEROL, ... | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of DynF from the Dynemicin Biosynthesis Pathway of Micromonospora chersina
To Be Published
|
|
6VZX
 
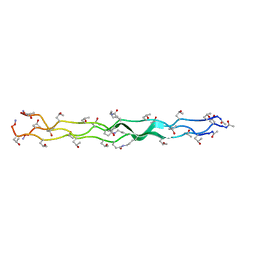 | | Structure of a Covalently Captured Collagen Triple Helix using Lysine-Glutamate Pairs | | Descriptor: | collagen mimetic peptide | | Authors: | Miller, M.D, Hulgan, S.A, Xu, W, Kosgei, A.J, Phillips Jr, G.N, Hartgerink, J.D. | | Deposit date: | 2020-02-28 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Covalent Capture of Collagen Triple Helices Using Lysine-Aspartate and Lysine-Glutamate Pairs.
Biomacromolecules, 21, 2020
|
|
3G5T
 
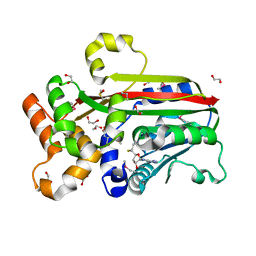 | | Crystal structure of trans-aconitate 3-methyltransferase from yeast | | Descriptor: | (2E)-2-(2-methoxy-2-oxoethyl)but-2-enedioic acid, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Burgie, E.S, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-03 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.119 Å) | | Cite: | Crystal structure of trans-aconitate 3-methyltransferase from yeast
To be Published
|
|
3GAN
 
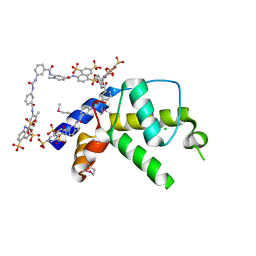 | | Crystal structure of gene product from Arabidopsis thaliana At3g22680 with bound suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, CHLORIDE ION, Uncharacterized protein At3g22680 | | Authors: | Burgie, E.S, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-02-17 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of gene product from Arabidopsis thaliana At3g22680 with bound suramin
To be Published
|
|
3H7C
 
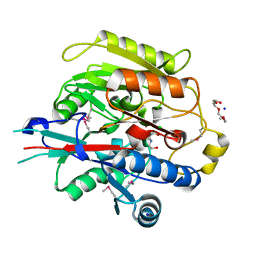 | | Crystal Structure of Arabidopsis thaliana Agmatine Deiminase from Cell Free Expression | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, Agmatine deiminase, CHLORIDE ION, ... | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Arabidopsis thaliana Agmatine Deiminase
To be Published
|
|
3IHR
 
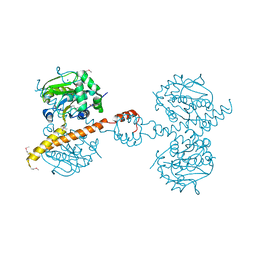 | | Crystal Structure of Uch37 | | Descriptor: | FORMIC ACID, SODIUM ION, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-11 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural characterization of human Uch37.
Proteins, 80, 2012
|
|
3H7K
 
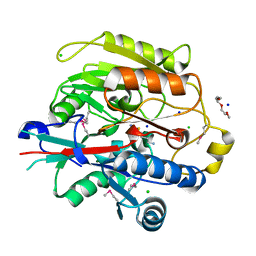 | |
1GFL
 
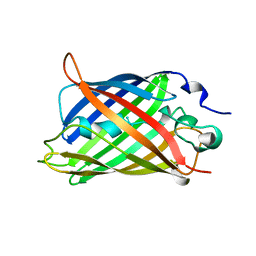 | |
6ND7
 
 | | The crystal structure of TerB co-crystallized with polyporic acid | | Descriptor: | 2~3~,2~6~-dihydroxy[1~1~,2~1~:2~4~,3~1~-terphenyl]-2~2~,2~5~-dione, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
6N04
 
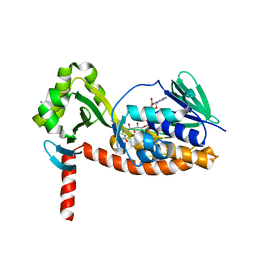 | | The X-ray crystal structure of AbsH3, an FAD dependent reductase from the Abyssomicin biosynthesis pathway in Streptomyces | | Descriptor: | AbsH3, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Clinger, J.A, Wang, X, Cai, W, Miller, M.D, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-11-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | The crystal structure of AbsH3: A putative flavin adenine dinucleotide-dependent reductase in the abyssomicin biosynthesis pathway.
Proteins, 2020
|
|
4TKT
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS6 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.4289 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4UC4
 
 | |
1MTK
 
 | |
1MTI
 
 | |
5CQF
 
 | | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae | | Descriptor: | IODIDE ION, L-lysine 6-monooxygenase | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Weerth, R.S, Cao, H, Yennamalli, R, Phillips Jr, G.N, Thomas, M.G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae
To Be Published
|
|
5DU2
 
 | | Structural analysis of EspG2 glycosyltransferase | | Descriptor: | EspG2 glycosyltransferase | | Authors: | Michalska, K, Elshahawi, S.I, Bigelow, L, Babnigg, G, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-09-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of EspG2 glycosyltransferase
To Be Published
|
|
5F4Z
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus | | Descriptor: | (1~{R},2~{R})-2,3-dihydro-1~{H}-indene-1,2-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, BABNIGG, G, BINGMAN, C.A, YENNAMALLI, R, LOHMAN, J, Chang, C.Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-12-03 | | Release date: | 2016-02-17 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus
To Be Published
|
|
5F1P
 
 | | Crystal Structure of Dehydrogenase from Streptomyces platensis | | Descriptor: | PtmO8 | | Authors: | Kim, Y, Li, H, Endres, M, Babnigg, G, Rudolf, J, Ma, M, Chang, C.-Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-11-30 | | Release date: | 2015-12-30 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal Structure of a Dehydrogenase, PtmO8, from Streptomyces platensis
To Be Published
|
|
