2FOB
 
 | | Structure of porcine pancreatic elastase in 40/50/10 cyclohexane | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
2FOC
 
 | | Structure of porcine pancreatic elastase in 55% dimethylformamide | | Descriptor: | CALCIUM ION, DIMETHYLFORMAMIDE, SULFATE ION, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
2FOA
 
 | | Structure of porcine pancreatic elastase in 40/50/10 % benzene | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
2FO9
 
 | | Structure of porcine pancreatic elastase in 95% acetone | | Descriptor: | ACETONE, CALCIUM ION, SULFATE ION, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
2FOG
 
 | | Structure of porcine pancreatic elastase in 40% trifluoroethanol | | Descriptor: | CALCIUM ION, SULFATE ION, TRIFLUOROETHANOL, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
2FOD
 
 | | Structure of porcine pancreatic elastase in 80% ethanol | | Descriptor: | CALCIUM ION, ETHANOL, SULFATE ION, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
2FOE
 
 | | Structure of porcine pancreatic elastase in 80% hexane | | Descriptor: | (2S)-HEX-5-ENE-1,2-DIOL, CALCIUM ION, SULFATE ION, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
2ETL
 
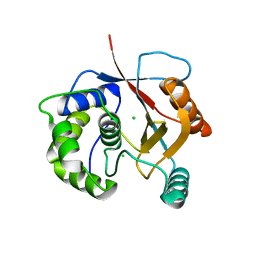 | | Crystal Structure of Ubiquitin Carboxy-terminal Hydrolase L1 (UCH-L1) | | Descriptor: | CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Das, C, Hoang, Q.Q, Kreinbring, C.A, Luchansky, S.J, Meray, R.K, Ray, S.S, Lansbury, P.T, Ringe, D, Petsko, G.A. | | Deposit date: | 2005-10-27 | | Release date: | 2006-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
5R42
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 7.5, room temperature | | Descriptor: | IODIDE ION, gamma-Chymotrypsin, peptide SWPW, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R49
 
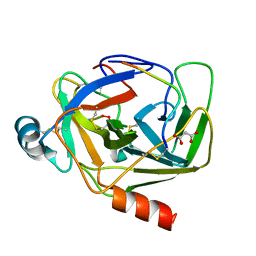 | | Crystal Structure of gamma-Chymotrypsin at pH 5.6, cryo temperature | | Descriptor: | IODIDE ION, MALONATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R45
 
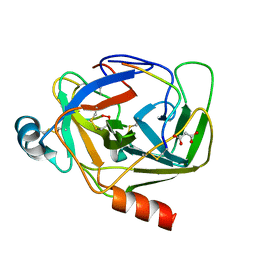 | | Crystal Structure of gamma-Chymotrypsin at pH 7.5, cryo temperature | | Descriptor: | Chymotrypsinogen A, IODIDE ION, MALONATE ION, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R4C
 
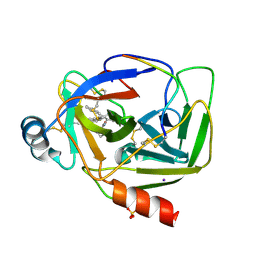 | | Crystal Structure of gamma-Chymotrypsin at pH 9, room temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R48
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 5.6, room temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R4A
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 9, room temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R44
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 7.5, room temperature | | Descriptor: | Chymotrypsinogen A, IODIDE ION, peptide SWPW, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R46
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 5.6, room temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R43
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 7.5, cryo temperature | | Descriptor: | Chymotrypsinogen A, IODIDE ION, MALONIC ACID, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R4B
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 9, cryo temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R4D
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 9, cryo temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R47
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 5.6, cryo temperature | | Descriptor: | IODIDE ION, MALONIC ACID, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
4PST
 
 | | Multiconformer model for Escherichia coli dihydrofolate reductase at 277 K | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | Deposit date: | 2014-03-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
4PTJ
 
 | | Ensemble model for Escherichia coli dihydrofolate reductase at 277K | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | Deposit date: | 2014-03-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
4PSS
 
 | | Multiconformer model for Escherichia coli dihydrofolate reductase at 100K | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | Deposit date: | 2014-03-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.849 Å) | | Cite: | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
4J5H
 
 | | Crystal Structure of B. thuringiensis AiiA mutant F107W with N-decanoyl-L-homoserine bound at the active site | | Descriptor: | GLYCEROL, N-acyl homoserine lactonase, N-decanoyl-L-homoserine, ... | | Authors: | Liu, C.F, Liu, D, Momb, J, Thomas, P.W, Lajoie, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2013-02-08 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A phenylalanine clamp controls substrate specificity in the quorum-quenching metallo-gamma-lactonase from Bacillus thuringiensis.
Biochemistry, 52, 2013
|
|
2ACU
 
 | | TYROSINE-48 IS THE PROTON DONOR AND HISTIDINE-110 DIRECTS SUBSTRATE STEREOCHEMICAL SELECTIVITY IN THE REDUCTION REACTION OF HUMAN ALDOSE REDUCTASE: ENZYME KINETICS AND THE CRYSTAL STRUCTURE OF THE Y48H MUTANT ENZYME | | Descriptor: | ALDOSE REDUCTASE, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bohren, K.M, Grimshaw, C.E, Lai, C.-J, Gabbay, K.H, Petsko, G.A, Harrison, D.H, Ringe, D. | | Deposit date: | 1994-04-15 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Tyrosine-48 is the proton donor and histidine-110 directs substrate stereochemical selectivity in the reduction reaction of human aldose reductase: enzyme kinetics and crystal structure of the Y48H mutant enzyme.
Biochemistry, 33, 1994
|
|
